Team:NYMU-Taipei/Project/Inhibition/Sensor
From 2013.igem.org
| (82 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NYMU-Taipei/Header}} | {{:Team:NYMU-Taipei/Header}} | ||
| + | |||
| + | =Sensing ''Nosema ceranae''= | ||
=='''Introduction'''== | =='''Introduction'''== | ||
| - | + | After ''Nosema ceranae'' enters the midgut of bees, we have to make ''E. coli'' sense this pathogen. ''E. coli'' with ROS-sensing promoters will respond to Reactive Oxygen Species (ROS) produced by infected midgut cells. The following circuit behind ROS-sensing promoters will be turned on and fight against ''N. cerenae''. | |
| - | + | ||
| - | + | For the circuit design, we added a new composition part: [http://parts.igem.org/Part:BBa_K1104250 device:BBa_K1104250] consisting of a constitutive promoter [http://parts.igem.org/Part:BBa_J23102 Part:BBa_J23102] and a transcription factor: OxyR ([http://parts.igem.org/Part:BBa_K1104200 Part:BBa_K1104200]), which can activate ROS-sensing promoters, increasing the quantity of inactivated transcription factors in order to enhance the response of promoters. Additionally, RBS ([http://parts.igem.org/Part:BBa_B0034 Part:BBa_B0034]) and double terminator [http://parts.igem.org/Part:BBa_B0015 Part:BBa_B0015]. Actually, the addition of [http://parts.igem.org/Part:BBa_K1104250 device:BBa_K1104250] remarkably enhanced the strength of promoter AhpCp2D1([http://parts.igem.org/Part:BBa_K1104204 Part:BBa_K1104204]) in our experiment results. | |
| + | |||
| + | We improved the function of a BioBrick Part: ahpC promoter ([http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001]) originally designed by [https://2010.igem.org/Team:KIT-Kyoto/Parts 2010 KIT-Tokyo team]. | ||
=='''Background''' == | =='''Background''' == | ||
===''Nosema Ceranae'' Induces Oxidative Stress in the Midgut.=== | ===''Nosema Ceranae'' Induces Oxidative Stress in the Midgut.=== | ||
| - | ROS(Reactive Oxygen Species)are efficient antimicrobial molecules. Genes involved in oxidation-reduction were significantly overrepresented in the gene set upregulated upon spore infection. In response to the infection, the increase of oxidoreduction in the midgut epithelia of bees infected by ''N. ceranae'' would therefore indicate an enhanced generation of ROS. It is suggested that ROS production is a general immune response in bees' midgut to microorganism infection. There are promoters in the genome of ''E. coli'' that can be regulated by ROS. After ''Nosema ceranae'' enter midgut of bee, the regulator of promoters will be activated by ROS, trigger the following circuit fighting against ''Nosema ceranae''. | + | ROS (Reactive Oxygen Species) are efficient antimicrobial molecules. Genes involved in oxidation-reduction were significantly overrepresented in the gene set upregulated upon spore infection. In response to the infection, the increase of oxidoreduction in the midgut epithelia of bees infected by ''N. ceranae'' would therefore indicate an enhanced generation of ROS. It is suggested that ROS production is a general immune response in bees' midgut to microorganism infection. There are promoters in the genome of ''E. coli'' that can be regulated by ROS. After ''Nosema ceranae'' enter midgut of bee, the regulator of promoters will be activated by ROS, trigger the following circuit fighting against ''Nosema ceranae''. |
===Literature Study of ROS-induced Promoters=== | ===Literature Study of ROS-induced Promoters=== | ||
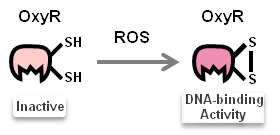
| + | Once ''N.cerenae'' infects the midgut of bees, midgut cells of bees will secrete Reactive Oxygen Species (ROS). ROS have been shown to be toxic but also function as signalling molecules which can transcriptionally activate transcription factors: OxyR ([http://parts.igem.org/Part:BBa_K1104200 Part:BBa_K1104200]). We focus on ROS-induced promoters regulated by OxyR. | ||
| + | [[File: NYMU_OxyRactive.png|frame|1000px|center|'''OxyR Activation'''<br>OxyR regulates the H<sub>2</sub>O<sub>2</sub> response of <br />''E. coli''. H<sub>2</sub>O<sub>2</sub> oxidizes OxyR Cys199 to an R-SOH that reacts with Cys208 in an intramolecular disulphide bond.]] | ||
| + | Activated transcription factor, OxyR, then binds to TFBS of promoters (sensor), as the pictures shown below: | ||
| + | {| | ||
| + | |[[File: NYMU_hemh.png|thumb|400px|center|'''[http://biocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU0-12951&orgids=ECOLI HemHp]([http://parts.igem.org/Part:BBa_K1104202 K1104202])''']] | ||
| + | |[[File: NYMU_trxc.png|thumb|490px|center|'''[http://biocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU0-3104&detail-level=1 TrxCp]([http://parts.igem.org/Part:BBa_K1104201 K1104201])''']] | ||
| + | |} | ||
| - | + | We did a study about promoters that can sense OxyR. Several promoters on the part registry can be induced by ROS: OxyR-activated promoters such as [http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001](ahpC)and [http://parts.igem.org/Part:BBa_K362005 Part:BBa_K362005](sufA)were designed and used as favorite parts by [https://2010.igem.org/Team:KIT-Kyoto/Parts 2010 KIT-Tokyo team]. They can be controlled by OxyR. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | We did a study about promoters that can sense OxyR | + | |
| - | We also got the | + | We also got the promoter sequences taken from [http://ecocyc.org/ Ecocyc] and tested promoters: [http://biocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU0-12951&orgids=ECOLI HemHp] and [http://biocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU0-3104&detail-level=1 TrxCp]. Both of them have dual binding site only interact with OxyR, cooperate with sigma factor 70, and have no E X S P cutting site. We have designed and sent them to PartRegistry as new parts: [http://parts.igem.org/Part:BBa_K1104202 Part:BBa_K1104202]、[http://parts.igem.org/Part:BBa_K1104201 Part:BBa_K1104201]. |
=='''Method'''== | =='''Method'''== | ||
==='''How Can ''Bee. coli'' Sense ''Nosema?'' '''=== | ==='''How Can ''Bee. coli'' Sense ''Nosema?'' '''=== | ||
| - | After ''Nosema ceranae'' infected midgut cells of bees, and | + | After ''Nosema ceranae'' infected midgut cells of bees, and ''Bee. coli'' should sense the pathogen first before the following circuit(fighting against ''Nosema ceranae'')is triggered. |
After ''Nosema ceranae'' infected midgut cells of bees, ROS(Reactive Oxygen Species) will be produced by midgut cells of bee and released to the cavity. As we can see, there are many kinds of promoters (and their transcription factors) responding to ROS naturally exist in ''E. coli MG1655'' genome. We choose proper and strength enough ones from them, basing on reporting assays we made about promoetr testing. Besides, in order to enhance the strength, we designed an upstream device to enlarge the expression of the activators, so the sensitivity of ''Bee. coli'' to ROS will be sharper, and the probability of downstream circuit(fighting against ''Nosema ceranae'') being turned on. | After ''Nosema ceranae'' infected midgut cells of bees, ROS(Reactive Oxygen Species) will be produced by midgut cells of bee and released to the cavity. As we can see, there are many kinds of promoters (and their transcription factors) responding to ROS naturally exist in ''E. coli MG1655'' genome. We choose proper and strength enough ones from them, basing on reporting assays we made about promoetr testing. Besides, in order to enhance the strength, we designed an upstream device to enlarge the expression of the activators, so the sensitivity of ''Bee. coli'' to ROS will be sharper, and the probability of downstream circuit(fighting against ''Nosema ceranae'') being turned on. | ||
==='''Circuit Design'''=== | ==='''Circuit Design'''=== | ||
| - | + | In order to enhance the strength and sensitivity of ROS-sensing promoters, we made ''E. coli'' continuously produce inactivated OxyR, so that ROS can affect promoters more easily. | |
| - | In order to enhance the strength and sensitivity of ROS-sensing promoters, we | + | |
====OxyR-included circuit==== | ====OxyR-included circuit==== | ||
| - | [[Image:NYMU_O12.png|frame|center|'''OxyR-included circuit(Device1+Device2)''']] | + | [[Image:NYMU_O12.png|frame|center|'''OxyR-included circuit(Device1+Device2)''']] |
| + | |||
| + | In OxyR-included circuit, Device2 is composed of sensor (OxyR-induced promoter, including TrxCp, HemHp, sufA, AhpCp1000, AhpCp2D1, AhpCp2, AhpCpD1, AhpCp1, DsbGp) plus reporter ([http://parts.igem.org/Part:BBa_E0840 Part:BBa_E0840]). | ||
| + | |||
| + | Device1 in order to enhance the effect of ROS on ''E. coli'' is added ahead: a constitutive promoter([http://parts.igem.org/Part:BBa_J23102 Part:BBa_J23102]), RBS([http://parts.igem.org/Part:BBa_B0034 Part:BBa_B0034]), activator OxyR([http://parts.igem.org/Part:BBa_K1104200 Part:BBa_K1104200]),and double terminator [http://parts.igem.org/Part:BBa_B0015 Part:BBa_B0015]. | ||
| + | |||
| + | Putting Device1 [http://parts.igem.org/Part:BBa_K1104250 device:BBa_K1104250] upstream to promoter AhpCp2D1([http://parts.igem.org/Part:BBa_K1104204 Part:BBa_K1104204]) remarkably enhanced the strength of promoter AhpCp2D1([http://parts.igem.org/Part:BBa_K1104204 Part:BBa_K1104204]) as our experiment results shows. | ||
| + | |||
| + | {| | ||
| + | |[[Image:NYMU_O1.png|frame|center|'''Device1 ([http://parts.igem.org/Part:BBa_K1104250 Part:BBa_K1104250])'''<br>a constitutive promoter([http://parts.igem.org/Part:BBa_J23102 Part:BBa_J23102])+ RBS([http://parts.igem.org/Part:BBa_B0034 Part:BBa_B0034])+ activator OxyR([http://parts.igem.org/Part:BBa_K1104200 Part:BBa_K1104200])+ double terminator [http://parts.igem.org/Part:BBa_B0015 Part:BBa_B0015]]] | ||
| + | |[[Image:NYMU_O2.png|frame|center|'''Device2'''<br>a sensor (OxyR-induced promoter, including TrxCp, HemHp, sufA, AhpCp1000, AhpCp2D1, AhpCp2, AhpCpD1, AhpCp1, DsbGp) + reporter ([http://parts.igem.org/Part:BBa_E0840 Part:BBa_E0840])]] | ||
| + | |} | ||
| - | |||
| - | |||
=====Parts===== | =====Parts===== | ||
{| | {| | ||
| Line 41: | Line 53: | ||
|[[File: NYMU_B0034.png |thumb|center|400px|'''RBS'''<br>([http://parts.igem.org/Part:BBa_B0034 Part:BBa_B0034])]] | |[[File: NYMU_B0034.png |thumb|center|400px|'''RBS'''<br>([http://parts.igem.org/Part:BBa_B0034 Part:BBa_B0034])]] | ||
|[[File: NYMU_OxyRcod.png |thumb|center|400px|'''OxyR coding'''<br>([http://parts.igem.org/Part:BBa_K1104200 Part:BBa_K1104200])]] | |[[File: NYMU_OxyRcod.png |thumb|center|400px|'''OxyR coding'''<br>([http://parts.igem.org/Part:BBa_K1104200 Part:BBa_K1104200])]] | ||
| + | |} | ||
| + | {| | ||
|[[File: NYMU_ B0015.png |thumb|center|400px|'''[http://parts.igem.org/Part:BBa_B0015 Part:BBa_B0015]''']] | |[[File: NYMU_ B0015.png |thumb|center|400px|'''[http://parts.igem.org/Part:BBa_B0015 Part:BBa_B0015]''']] | ||
|[[File:NYMU_E0840.png|thumb|center|400px|'''[http://parts.igem.org/Part:BBa_E0840 Part:BBa_E0840]''']] | |[[File:NYMU_E0840.png|thumb|center|400px|'''[http://parts.igem.org/Part:BBa_E0840 Part:BBa_E0840]''']] | ||
| Line 48: | Line 62: | ||
We searched for promoters that can be induced with ROS. Most of them activated by OxyR. | We searched for promoters that can be induced with ROS. Most of them activated by OxyR. | ||
*Promoters on Ecocyc | *Promoters on Ecocyc | ||
| - | [[File:NYMU_ TrxC.png|thumb|center|400px|''' | + | {| |
| - | [[File:NYMU_ HemH.png|thumb|center|400px|''' | + | |[[File:NYMU_ TrxC.png|thumb|center|400px|''' TrxCp '''<br>([http://parts.igem.org/Part:BBa_K1104201 Part:BBa_K1104201])]] |
| + | |[[File:NYMU_ HemH.png|thumb|center|400px|''' HemHp '''<br>([http://parts.igem.org/Part:BBa_K1104202 Part:BBa_K1104202])]] | ||
| + | |} | ||
*Promoters on PartRegistry | *Promoters on PartRegistry | ||
| - | [[File:NYMU_ SufA.png|thumb|center|400px|''' | + | [[File:NYMU_ SufA.png|thumb|center|400px|''' sufA '''<br>([http://parts.igem.org/Part:BBa_K362005 Part:BBa_K362005])]] |
*Improved-Promoters from [http://parts.igem.org/Part:BBa_K362001 ahpC(Part:BBa_K362001)] | *Improved-Promoters from [http://parts.igem.org/Part:BBa_K362001 ahpC(Part:BBa_K362001)] | ||
| - | [[File:NYMU_Amutate1000.png| | + | {| |
| - | [[File:NYMU_AhpC2D1.png|thumb|center|400px|''' | + | |[[File:NYMU_Amutate1000.png|frame|center|''' AhpCp1000'''[http://parts.igem.org/Part:BBa_K1104203 Part:BBa_K1104203]]] |
| - | + | |} | |
| - | [[File:NYMU_AhpCD1.png|thumb|center|400px|''' | + | {| |
| - | + | |[[File:NYMU_AhpC2D1.png|thumb|center|400px|''' AhpCp2D1'''<br>([http://parts.igem.org/Part:BBa_K1104204 Part:BBa_K1104204])]] | |
| - | [[File: | + | |[[File:NYMU_AhpCD1.png|thumb|center|400px|''' AhpCpD1'''<br>([http://parts.igem.org/Part:BBa_K1104206 Part:BBa_K1104206])]] |
| - | + | |} | |
| - | + | {| | |
| - | + | |[[File:NYMU_AhpC1.png|thumb|center|400px|''' AhpCp1'''<br>([http://parts.igem.org/Part:BBa_K1104207 Part:BBa_K1104207])]] | |
| - | + | |[[File:NYMU_AhpC2.png|thumb|center|400px|''' AhpCp2'''<br>([http://parts.igem.org/Part:BBa_K1104205 Part:BBa_K1104205])]] | |
| - | + | |[[File:NYMU_DsbG.png|thumb|center|400px|''' DsbGp'''<br>([http://parts.igem.org/Part:BBa_K1104207 Part:BBa_K1104207])]] | |
| - | [[File: | + | |} |
| - | + | ||
| - | [[File: | + | |
| - | + | ||
| - | + | ||
===Disscussion=== | ===Disscussion=== | ||
| - | We | + | We tested Device2 of each OxyR-induced promoters, as well as Device1+Device2 of AhpCp2D1. And the results of OxyR-induced promoters testing on PartRegistry are linked as follows:<br> |
| - | + | *TrxCp ([http://parts.igem.org/Part:BBa_K1104201:Experience Part:BBa_K1104201:Experience]) | |
| - | + | *HemHp ([http://parts.igem.org/Part:BBa_K1104202:Experience Part:BBa_K1104202:Experience]) | |
| - | + | *sufAp ([http://parts.igem.org/Part:BBa_K1104205:Experience Part:BBa_K1104205:Experience]) | |
| - | + | *AhpCp1000 ([http://parts.igem.org/Part:BBa_K1104203:Experience Part:BBa_K1104203:Experience]) | |
| - | + | *AhpCp2D1 ([http://parts.igem.org/Part:BBa_K1104204:Experience Part:BBa_K1104204:Experience]) | |
| - | + | *AhpCp2 ([http://parts.igem.org/Part:BBa_K1104205:Experience Part:BBa_K1104205:Experience]) | |
| - | + | *AhpCpD1 ([http://parts.igem.org/Part:BBa_K1104206:Experience Part:BBa_K1104206:Experience]) | |
| - | + | *AhpCp1 ([http://parts.igem.org/Part:BBa_K1104207:Experience Part:BBa_K1104207:Experience]) | |
| - | + | *DsbGp ([http://parts.igem.org/Part:BBa_K1104208:Experience Part:BBa_K1104208:Experience]) | |
| - | + | ||
| + | [[File:NYMU pp.png|frame|center]]Above shows the strengths of the basal level of various promoters that respond to reactive oxygen species with respect to BBa_J23101. | ||
| + | |||
| + | [[Image:NYMU_2d1.png|frame|center|In the presence of Device 1, AhpCp2D1 shows 7 to 9 folds of strength (Error bars represent the 95% confidence interval)]] | ||
==='''Part Improvement'''=== | ==='''Part Improvement'''=== | ||
| + | [[File:NYMU_originalA.png|frame|center|'''ahpC([http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001])'''<br>There is a Pst1 site in the red rectangle.]] | ||
| + | We improved the function of a BioBrick Part: ahpC promoter ([http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001]) designed by [https://2010.igem.org/Team:KIT-Kyoto/Parts 2010 KIT-Tokyo team] into four versions. On the PartRegistry, the complex part according to [http://ecocyc.org/ECOLI/new-image?object=EG11384 Ecocyc] composition contains part of DsbG coding, hybrid promoters, shared TFBS (Transcription Factor Binding Site), and reverse promoter DsbGp. The intergenic region between DsbGp and AhpCp1 carries two binding sites for OxyR. | ||
| - | + | [[File:NYMU_Amutate1000.png|thumb|600px|center|'''AhpCp1000[http://parts.igem.org/Part:BBa_K1104203 Part:BBa_K1104203]'''<br>We succesfully mutated the Pst1 site (ctgcag->ctacag) of K362001.]] | |
| + | {| | ||
| + | |[[File:NYMU_AhpC2D1.png|thumb|400px|center|'''AhpCp2D1([http://parts.igem.org/Part:BBa_K1104204 Part:BBa_K1104204])'''<br>We strimmed DsbG coding out of the original sequence, making the functional sequence shorter and more efficient.]] | ||
| + | |[[File:NYMU_AhpCD1.png|thumb|400px|center|'''AhpCpD1([http://parts.igem.org/Part:BBa_K1104206 Part:BBa_K1104206])'''<br>We designed a dual-side hybrid promoter, including AhpCp1 and DsbGp(reverse), and two binding sites for OxyR. OxyR binding to TFBS leads to bidirectional transcription.]] | ||
| + | |} | ||
| - | + | Because ahpC ([http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001]) is a hybrid promoter containing AhpCp1, AhpCp2, and DsbGp promoters in its 1000bp, we created the new part and test it apart. | |
| - | [[File: | + | {| |
| - | [[File: | + | |[[File:NYMU_AhpC1.png|thumb|200px|center|'''AhpCp1'''<br>[http://parts.igem.org/Part:BBa_K1104207 Part:BBa_K1104207]]] |
| - | [[File: | + | |[[File:NYMU_AhpC2.png|thumb|200px|center|'''AhpCp2'''<br>[http://parts.igem.org/Part:BBa_K1104205 Part:BBa_K1104205]]] |
| + | |[[File:NYMU_DsbG.png|thumb|center|400px|'''DsbGp'''<br>([http://parts.igem.org/Part:BBa_K1104207 Part:BBa_K1104207])]] | ||
| + | |} | ||
| - | + | ===Discussion=== | |
| - | + | As the results show, we successfully improved ahpC promoter ([http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001]). And the results and disscussion on the PartRegistry are linked as follows:<br> | |
| - | [ | + | *AhpCp1000([http://parts.igem.org/Part:BBa_K1104203:Experience Part:BBa_K1104203:Experience]) |
| + | *AhpCp2D1([http://parts.igem.org/Part:BBa_K1104204:Experience Part:BBa_K1104204:Experience]) | ||
| + | *AhpCp2([http://parts.igem.org/Part:BBa_K1104205:Experience Part:BBa_K1104205:Experience]) | ||
| + | *AhpCpD1([http://parts.igem.org/Part:BBa_K1104206:Experience Part:BBa_K1104206:Experience]) | ||
| + | *AhpCp1([http://parts.igem.org/Part:BBa_K1104207:Experience Part:BBa_K1104207:Experience]) | ||
| + | *DsbGp([http://parts.igem.org/Part:BBa_K1104208:Experience Part:BBa_K1104208:Experience]) | ||
| - | + | [[File: NYMU_AA.png|frame|center]]Above shows promoter testing of ahpC promoter [http://parts.igem.org/Part:BBa_K362001 Part:K362001]) and its improvements, which suggests that AhpCpd1 and AhpCp1 show so weak promoter strength that we only focus on the change of promoter strength of the three promoters as following: | |
| - | + | [[File: NYMU_AAAA.png|frame|center]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==''' | + | =='''References'''== |
| - | + | *Dussaubat, Claudia, Brunet, Jean-Luc, Higes, Mariano, Colbourne, John K., Lopez, Jacqueline, Choi, Jeong-Hyeon, Martín-Hernández, Raquel, ... Alaux, Cédric. (n.d.). Gut Pathology and Responses to the Microsporidium Nosema ceranae in the Honey Bee Apis mellifera. Public Library of Science. | |
| - | + | *D'Autréaux, B., & Toledano, M. B. (January 01, 2007). ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nature Reviews. Molecular Cell Biology, 8, 10, 813-24.2[http://www.nature.com/nrm/journal/v8/n10/fig_tab/nrm2256_F1.html] | |
| - | + | *Ryu, S. E. (June 01, 2012). Structural mechanism of disulphide bond-mediated redox switches. Journal of Biochemistry, 151, 6, 579-588.[http://jb.oxfordjournals.org/content/early/2012/05/02/jb.mvs046.full.pdf] | |
| - | + | *[http://ecocyc.org/ Ecocyc]. | |
Latest revision as of 05:27, 28 October 2013


Contents |
Sensing Nosema ceranae
Introduction
After Nosema ceranae enters the midgut of bees, we have to make E. coli sense this pathogen. E. coli with ROS-sensing promoters will respond to Reactive Oxygen Species (ROS) produced by infected midgut cells. The following circuit behind ROS-sensing promoters will be turned on and fight against N. cerenae.
For the circuit design, we added a new composition part: [http://parts.igem.org/Part:BBa_K1104250 device:BBa_K1104250] consisting of a constitutive promoter [http://parts.igem.org/Part:BBa_J23102 Part:BBa_J23102] and a transcription factor: OxyR ([http://parts.igem.org/Part:BBa_K1104200 Part:BBa_K1104200]), which can activate ROS-sensing promoters, increasing the quantity of inactivated transcription factors in order to enhance the response of promoters. Additionally, RBS ([http://parts.igem.org/Part:BBa_B0034 Part:BBa_B0034]) and double terminator [http://parts.igem.org/Part:BBa_B0015 Part:BBa_B0015]. Actually, the addition of [http://parts.igem.org/Part:BBa_K1104250 device:BBa_K1104250] remarkably enhanced the strength of promoter AhpCp2D1([http://parts.igem.org/Part:BBa_K1104204 Part:BBa_K1104204]) in our experiment results.
We improved the function of a BioBrick Part: ahpC promoter ([http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001]) originally designed by 2010 KIT-Tokyo team.
Background
Nosema Ceranae Induces Oxidative Stress in the Midgut.
ROS (Reactive Oxygen Species) are efficient antimicrobial molecules. Genes involved in oxidation-reduction were significantly overrepresented in the gene set upregulated upon spore infection. In response to the infection, the increase of oxidoreduction in the midgut epithelia of bees infected by N. ceranae would therefore indicate an enhanced generation of ROS. It is suggested that ROS production is a general immune response in bees' midgut to microorganism infection. There are promoters in the genome of E. coli that can be regulated by ROS. After Nosema ceranae enter midgut of bee, the regulator of promoters will be activated by ROS, trigger the following circuit fighting against Nosema ceranae.
Literature Study of ROS-induced Promoters
Once N.cerenae infects the midgut of bees, midgut cells of bees will secrete Reactive Oxygen Species (ROS). ROS have been shown to be toxic but also function as signalling molecules which can transcriptionally activate transcription factors: OxyR ([http://parts.igem.org/Part:BBa_K1104200 Part:BBa_K1104200]). We focus on ROS-induced promoters regulated by OxyR.
Activated transcription factor, OxyR, then binds to TFBS of promoters (sensor), as the pictures shown below:
We did a study about promoters that can sense OxyR. Several promoters on the part registry can be induced by ROS: OxyR-activated promoters such as [http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001](ahpC)and [http://parts.igem.org/Part:BBa_K362005 Part:BBa_K362005](sufA)were designed and used as favorite parts by 2010 KIT-Tokyo team. They can be controlled by OxyR.
We also got the promoter sequences taken from [http://ecocyc.org/ Ecocyc] and tested promoters: [http://biocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU0-12951&orgids=ECOLI HemHp] and [http://biocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU0-3104&detail-level=1 TrxCp]. Both of them have dual binding site only interact with OxyR, cooperate with sigma factor 70, and have no E X S P cutting site. We have designed and sent them to PartRegistry as new parts: [http://parts.igem.org/Part:BBa_K1104202 Part:BBa_K1104202]、[http://parts.igem.org/Part:BBa_K1104201 Part:BBa_K1104201].
Method
How Can Bee. coli Sense Nosema?
After Nosema ceranae infected midgut cells of bees, and Bee. coli should sense the pathogen first before the following circuit(fighting against Nosema ceranae)is triggered. After Nosema ceranae infected midgut cells of bees, ROS(Reactive Oxygen Species) will be produced by midgut cells of bee and released to the cavity. As we can see, there are many kinds of promoters (and their transcription factors) responding to ROS naturally exist in E. coli MG1655 genome. We choose proper and strength enough ones from them, basing on reporting assays we made about promoetr testing. Besides, in order to enhance the strength, we designed an upstream device to enlarge the expression of the activators, so the sensitivity of Bee. coli to ROS will be sharper, and the probability of downstream circuit(fighting against Nosema ceranae) being turned on.
Circuit Design
In order to enhance the strength and sensitivity of ROS-sensing promoters, we made E. coli continuously produce inactivated OxyR, so that ROS can affect promoters more easily.
OxyR-included circuit
In OxyR-included circuit, Device2 is composed of sensor (OxyR-induced promoter, including TrxCp, HemHp, sufA, AhpCp1000, AhpCp2D1, AhpCp2, AhpCpD1, AhpCp1, DsbGp) plus reporter ([http://parts.igem.org/Part:BBa_E0840 Part:BBa_E0840]).
Device1 in order to enhance the effect of ROS on E. coli is added ahead: a constitutive promoter([http://parts.igem.org/Part:BBa_J23102 Part:BBa_J23102]), RBS([http://parts.igem.org/Part:BBa_B0034 Part:BBa_B0034]), activator OxyR([http://parts.igem.org/Part:BBa_K1104200 Part:BBa_K1104200]),and double terminator [http://parts.igem.org/Part:BBa_B0015 Part:BBa_B0015].
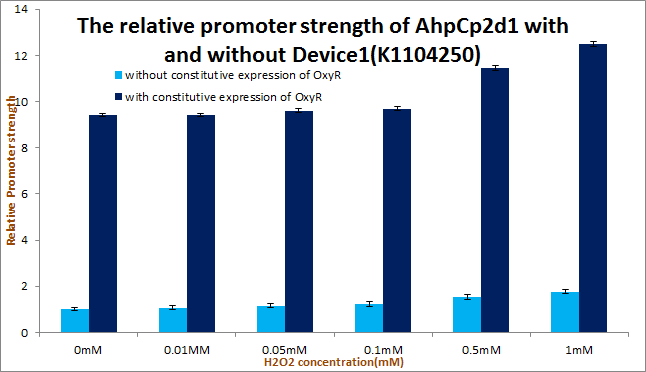
Putting Device1 [http://parts.igem.org/Part:BBa_K1104250 device:BBa_K1104250] upstream to promoter AhpCp2D1([http://parts.igem.org/Part:BBa_K1104204 Part:BBa_K1104204]) remarkably enhanced the strength of promoter AhpCp2D1([http://parts.igem.org/Part:BBa_K1104204 Part:BBa_K1104204]) as our experiment results shows.
Parts
OxyR-activated promoters
We searched for promoters that can be induced with ROS. Most of them activated by OxyR.
- Promoters on Ecocyc
- Promoters on PartRegistry
- Improved-Promoters from [http://parts.igem.org/Part:BBa_K362001 ahpC(Part:BBa_K362001)]
Disscussion
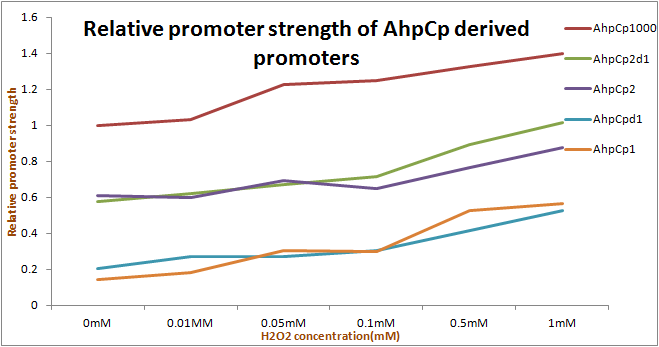
We tested Device2 of each OxyR-induced promoters, as well as Device1+Device2 of AhpCp2D1. And the results of OxyR-induced promoters testing on PartRegistry are linked as follows:
- TrxCp ([http://parts.igem.org/Part:BBa_K1104201:Experience Part:BBa_K1104201:Experience])
- HemHp ([http://parts.igem.org/Part:BBa_K1104202:Experience Part:BBa_K1104202:Experience])
- sufAp ([http://parts.igem.org/Part:BBa_K1104205:Experience Part:BBa_K1104205:Experience])
- AhpCp1000 ([http://parts.igem.org/Part:BBa_K1104203:Experience Part:BBa_K1104203:Experience])
- AhpCp2D1 ([http://parts.igem.org/Part:BBa_K1104204:Experience Part:BBa_K1104204:Experience])
- AhpCp2 ([http://parts.igem.org/Part:BBa_K1104205:Experience Part:BBa_K1104205:Experience])
- AhpCpD1 ([http://parts.igem.org/Part:BBa_K1104206:Experience Part:BBa_K1104206:Experience])
- AhpCp1 ([http://parts.igem.org/Part:BBa_K1104207:Experience Part:BBa_K1104207:Experience])
- DsbGp ([http://parts.igem.org/Part:BBa_K1104208:Experience Part:BBa_K1104208:Experience])
Part Improvement
We improved the function of a BioBrick Part: ahpC promoter ([http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001]) designed by 2010 KIT-Tokyo team into four versions. On the PartRegistry, the complex part according to [http://ecocyc.org/ECOLI/new-image?object=EG11384 Ecocyc] composition contains part of DsbG coding, hybrid promoters, shared TFBS (Transcription Factor Binding Site), and reverse promoter DsbGp. The intergenic region between DsbGp and AhpCp1 carries two binding sites for OxyR.
Because ahpC ([http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001]) is a hybrid promoter containing AhpCp1, AhpCp2, and DsbGp promoters in its 1000bp, we created the new part and test it apart.
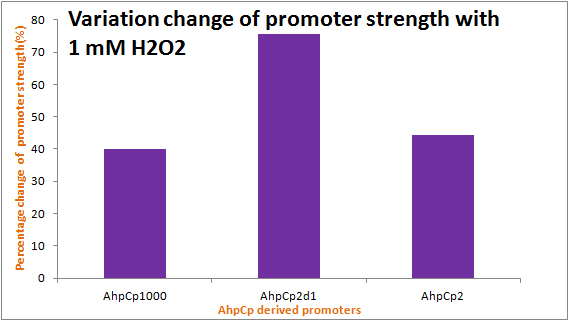
Discussion
As the results show, we successfully improved ahpC promoter ([http://parts.igem.org/Part:BBa_K362001 Part:BBa_K362001]). And the results and disscussion on the PartRegistry are linked as follows:
- AhpCp1000([http://parts.igem.org/Part:BBa_K1104203:Experience Part:BBa_K1104203:Experience])
- AhpCp2D1([http://parts.igem.org/Part:BBa_K1104204:Experience Part:BBa_K1104204:Experience])
- AhpCp2([http://parts.igem.org/Part:BBa_K1104205:Experience Part:BBa_K1104205:Experience])
- AhpCpD1([http://parts.igem.org/Part:BBa_K1104206:Experience Part:BBa_K1104206:Experience])
- AhpCp1([http://parts.igem.org/Part:BBa_K1104207:Experience Part:BBa_K1104207:Experience])
- DsbGp([http://parts.igem.org/Part:BBa_K1104208:Experience Part:BBa_K1104208:Experience])
References
- Dussaubat, Claudia, Brunet, Jean-Luc, Higes, Mariano, Colbourne, John K., Lopez, Jacqueline, Choi, Jeong-Hyeon, Martín-Hernández, Raquel, ... Alaux, Cédric. (n.d.). Gut Pathology and Responses to the Microsporidium Nosema ceranae in the Honey Bee Apis mellifera. Public Library of Science.
- D'Autréaux, B., & Toledano, M. B. (January 01, 2007). ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nature Reviews. Molecular Cell Biology, 8, 10, 813-24.2[http://www.nature.com/nrm/journal/v8/n10/fig_tab/nrm2256_F1.html]
- Ryu, S. E. (June 01, 2012). Structural mechanism of disulphide bond-mediated redox switches. Journal of Biochemistry, 151, 6, 579-588.[http://jb.oxfordjournals.org/content/early/2012/05/02/jb.mvs046.full.pdf]
- [http://ecocyc.org/ Ecocyc].
 "
"