Team:Shenzhen BGIC 0101/Requirements
From 2013.igem.org
| (33 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:Shenzhen BGIC 0101/Templates/Header}} | ||
<html> | <html> | ||
| - | < | + | <body> |
| - | + | <div id="HumanPractices"> | |
| - | + | <br/><h1>Human Practices</h1><br/> | |
| - | + | <h2> 1. <a href="https://2013.igem.org/Team:Shenzhen_BGIC_0101/Web-based_trial">Share Web-based server</a></h2><br/> | |
| - | + | <h2>2. Regional workshop</h2> | |
| - | + | <p>Regional workshop was held in BGI on May, 26th, 2013. As host, we invited 6 teams, SYSU-China, SYSU-Software, SCUT, SCAU-China, SUSTC-Shenzhen-A, SUSTC-Shenzhen-B. <br/> | |
| - | + | Each team shared their ideas and progress, and even established cooperative intention:<br/> | |
| - | + | DNA material shares among teams, SUSTC-Shenzhen-A assisted BGIC_Shenzhen_ATCG to purchase experimental material.<br/> | |
| - | + | You can find the news <a href=" http://www.genomics.cn/news/show_news?nid=99539">here</a></p> | |
| - | + | <br/> | |
| + | <h2>3. Talk with Dr. Patrick Yizhi Cai</h2> | ||
| + | <p>Dr. Patrick Cai is the Autodesk distinguished scholar and a Principal Investigator (with Chancellor's Fellowship), who directs the group of synthetic genomics in the university of Edinburgh. His research focuses on neochromosome design, computer assisted design for synbio and large scale DNA synthesis automation. So he is the most authoritative scholar for Genovo.<br/> | ||
| + | He came to BGI on Sep, 26th, 2013 and talk with us about Genovo's design and application. | ||
| + | "Genovo is quite a neat idea." Quoted as Cai.</p><br/> | ||
| + | <h2>4. T-shirt sale</h2><br/> | ||
| + | <p>We tried to sell our design iGEM shirt to BGI’employees.</p><br/> | ||
| + | <p>You may also order your own one <a href="https://docs.google.com/forms/d/1v8vvGN4TRq1yzLuKNed43mgl1KEyaWpOjh9VJfyNZqM/viewform">here</a>, if you’re in China.<br/> | ||
| + | </p> | ||
| + | </div> | ||
| + | <br/><br/> | ||
| + | <hr style="color:#7380AE; height:2px;" /> | ||
| - | + | <div id="Safety"> | |
| - | + | <br/><h1>Safety</h1><br/> | |
| - | + | <h3><img src="https://static.igem.org/mediawiki/2013/c/c0/Quest.png" width="30px" height="30px"/>Question1: Are you using the iGEM Software repository at github.com/igemsoftware?If you have instead stored your code elsewhere, please explain where and why you have put it there.If your code is not in the iGEM repository, are you using any version control system such as Git, CVS, or SVN?</h3> | |
| - | + | <p><b><img src="https://static.igem.org/mediawiki/2013/e/ef/Answer.png" width="30px" height="30px"/>Answer:Yes. </b></p> | |
| - | + | <h3><img src="https://static.igem.org/mediawiki/2013/c/c0/Quest.png" width="30px" height="30px"/>Question2:a. Does your software store any private data supplied by the user? (For example: the user's name and email address, passwords, DNA sequences, circuit designs, etc.) If yes, please describe what kind of data is stored. If no, skip the rest of this question and move on to question 3.</h3> | |
| - | + | <p><b><img src="https://static.igem.org/mediawiki/2013/e/ef/Answer.png" width="30px" height="30px"/>Answer:Yes. We store DNA sequences and circuit designs provided by users.</b></p> | |
| + | <h3><img src="https://static.igem.org/mediawiki/2013/c/c0/Quest.png" width="30px" height="30px"/>Question2:b. What is the URL or IP address where the user's private data is stored? Where is the physical computer or hard drive that contains the user's private data?</h3> | ||
| + | <p><b><img src="https://static.igem.org/mediawiki/2013/e/ef/Answer.png" width="30px" height="30px"/>Answer:The server is still being constructed. </b></p> | ||
| + | <h3><img src="https://static.igem.org/mediawiki/2013/c/c0/Quest.png" width="30px" height="30px"/>Question2:c. Please describe any encryption, password protection, etc. that you use to protect the user's data. (It is not mandatory to have such protections, but if you do, describe them.)</h3> | ||
| + | <p><b><img src="https://static.igem.org/mediawiki/2013/e/ef/Answer.png" width="30px" height="30px"/>Answer:No.</b></p> | ||
| + | <h3><img src="https://static.igem.org/mediawiki/2013/c/c0/Quest.png" width="30px" height="30px"/>Question3:Does your software include any other security features? Please describe them here.</h3> | ||
| + | <p><b><img src="https://static.igem.org/mediawiki/2013/e/ef/Answer.png" width="30px" height="30px"/>Answer:Yes. The software can store the data provided by users and those produced step by step, so that user won't miss their result.</b></p> | ||
| + | <h3><img src="https://static.igem.org/mediawiki/2013/c/c0/Quest.png" width="30px" height="30px"/>Question4:Does your software let the user create a design by choosing parts/genes from a list/database, such as the Registry? If so, which lists/databases are included? Is there any restriction on which parts/genes the user can choose?</h3> | ||
| + | <p><b><img src="https://static.igem.org/mediawiki/2013/e/ef/Answer.png" width="30px" height="30px"/>Answer:Yes. KEGG, partsregistry, Codon usage database, SGD, VFDB, E. coli Genome Project. We have no restriction on genes the user can choose.</b></p> | ||
| + | <h3><img src="https://static.igem.org/mediawiki/2013/c/c0/Quest.png" width="30px" height="30px"/>Question5:Does your software allow users to write new data into any public lists or databases? If so, do you check the new data for errors before allowing it to be written?</h3> | ||
| + | <p><b><img src="https://static.igem.org/mediawiki/2013/e/ef/Answer.png" width="30px" height="30px"/>Answer:No.</b></p> | ||
| + | <h3><img src="https://static.igem.org/mediawiki/2013/c/c0/Quest.png" width="30px" height="30px"/>Question6:Does your software include any other features that encourage the user to create safe designs? Please describe them here.</h3> | ||
| + | <p><b><img src="https://static.igem.org/mediawiki/2013/e/ef/Answer.png" width="30px" height="30px"/>Answer:Yes. We develop a plugin to show the relationship between the gene or artificial logic gate gene circuit to guide user to select their target genes safely. Besides, we have reminder about biosafely selection for users.</b></p> | ||
| + | <h3><img src="https://static.igem.org/mediawiki/2013/c/c0/Quest.png" width="30px" height="30px"/>Question7:Is your team also doing biological work in a wet lab?</h3> | ||
| + | <p><b><img src="https://static.igem.org/mediawiki/2013/e/ef/Answer.png" width="30px" height="30px"/>Answer:No.</b></p> | ||
| + | </div></article> | ||
| + | </section> | ||
| + | <br/><br/> | ||
| + | <hr style="color:#7380AE; height:2px;" /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | <div id="collaboration"> | |
| - | + | <br/><h1>Collaboration</h1><br/> | |
| - | < | + | <h2>Cooperation: Shenzhen_BGIC_ATCG</h2> |
| - | < | + | <br/> |
| - | + | <p>We use NucleoMod to assist other wet team, Shenzhen_BGIC_ATCG, to design the biobrick optimized for eukaryons. <br/> | |
| - | < | + | Below is one of the examples:<br/> |
| - | + | We transform the prokaryon version of cas9 for S.cerevisiae automatically.<br/> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/e/e4/Ww1.png" /><br/><br/> | |
| - | + | Firstly, delete five enzymes of biobrick standard, EcoRI, XbaI, SpeI, PstI, NotI based on synonymous mutation of CDS.<br/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | + | ||
| - | + | <img src="https://static.igem.org/mediawiki/2013/7/71/%E5%9B%BE%E7%89%872.png" width="100%" /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br/> | <br/> | ||
| - | + | Secondly, optimize the sequence for S.cerevisiae according to codon usage <a href="http://www.kazusa.or.jp/codon/ ">database</a>.<br/> | |
| - | + | Finally, we recreate the sequence and annotation file.<br/> | |
| - | + | <br/><br/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <h2>We tried out SegmMan module to automatically split designed yeast_chr07_4_51 into 2k minichunks in command line.</h2><br/> | |
| - | + | <p> | |
| - | </ | + | <img src="https://static.igem.org/mediawiki/2013/0/05/%E5%9B%BE%E7%89%873.png" /><br/> |
| - | < | + | After run the flow shell, SegmMan analyze the exited enzyme site along the sequence of yeast_chr07_4_51, and finally splited it into about 2k minichunks, so that we can synthesize these fragments and assemble them into whole.<br/> |
| - | < | + | The following is the final result of segmentations. yeast_chr07_4_51 is split into many 2k minichunks. After these fragments are synthesized, they can be assembled into whole step by step, from 2k to 10k to 30k to whole, according Gibson, Goldengate, Telomere Assembly.<br/> |
| - | + | On 2k and 10k level, they also have vectors homologous region and designed enzyme site, so that they can be preserved and cut out to use quickly. <br/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</p> | </p> | ||
| - | + | </div> | |
| - | + | <br/><br/> | |
| - | + | <hr style="color:#7380AE; height:2px;" /> | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</body> | </body> | ||
</html> | </html> | ||
| - | |||
Latest revision as of 20:52, 28 October 2013
Human Practices
1. Share Web-based server
2. Regional workshop
Regional workshop was held in BGI on May, 26th, 2013. As host, we invited 6 teams, SYSU-China, SYSU-Software, SCUT, SCAU-China, SUSTC-Shenzhen-A, SUSTC-Shenzhen-B.
Each team shared their ideas and progress, and even established cooperative intention:
DNA material shares among teams, SUSTC-Shenzhen-A assisted BGIC_Shenzhen_ATCG to purchase experimental material.
You can find the news here
3. Talk with Dr. Patrick Yizhi Cai
Dr. Patrick Cai is the Autodesk distinguished scholar and a Principal Investigator (with Chancellor's Fellowship), who directs the group of synthetic genomics in the university of Edinburgh. His research focuses on neochromosome design, computer assisted design for synbio and large scale DNA synthesis automation. So he is the most authoritative scholar for Genovo.
He came to BGI on Sep, 26th, 2013 and talk with us about Genovo's design and application.
"Genovo is quite a neat idea." Quoted as Cai.
4. T-shirt sale
We tried to sell our design iGEM shirt to BGI’employees.
You may also order your own one here, if you’re in China.
Safety
 Question1: Are you using the iGEM Software repository at github.com/igemsoftware?If you have instead stored your code elsewhere, please explain where and why you have put it there.If your code is not in the iGEM repository, are you using any version control system such as Git, CVS, or SVN?
Question1: Are you using the iGEM Software repository at github.com/igemsoftware?If you have instead stored your code elsewhere, please explain where and why you have put it there.If your code is not in the iGEM repository, are you using any version control system such as Git, CVS, or SVN?
 Answer:Yes.
Answer:Yes.
 Question2:a. Does your software store any private data supplied by the user? (For example: the user's name and email address, passwords, DNA sequences, circuit designs, etc.) If yes, please describe what kind of data is stored. If no, skip the rest of this question and move on to question 3.
Question2:a. Does your software store any private data supplied by the user? (For example: the user's name and email address, passwords, DNA sequences, circuit designs, etc.) If yes, please describe what kind of data is stored. If no, skip the rest of this question and move on to question 3.
 Answer:Yes. We store DNA sequences and circuit designs provided by users.
Answer:Yes. We store DNA sequences and circuit designs provided by users.
 Question2:b. What is the URL or IP address where the user's private data is stored? Where is the physical computer or hard drive that contains the user's private data?
Question2:b. What is the URL or IP address where the user's private data is stored? Where is the physical computer or hard drive that contains the user's private data?
 Answer:The server is still being constructed.
Answer:The server is still being constructed.
 Question2:c. Please describe any encryption, password protection, etc. that you use to protect the user's data. (It is not mandatory to have such protections, but if you do, describe them.)
Question2:c. Please describe any encryption, password protection, etc. that you use to protect the user's data. (It is not mandatory to have such protections, but if you do, describe them.)
 Answer:No.
Answer:No.
 Question3:Does your software include any other security features? Please describe them here.
Question3:Does your software include any other security features? Please describe them here.
 Answer:Yes. The software can store the data provided by users and those produced step by step, so that user won't miss their result.
Answer:Yes. The software can store the data provided by users and those produced step by step, so that user won't miss their result.
 Question4:Does your software let the user create a design by choosing parts/genes from a list/database, such as the Registry? If so, which lists/databases are included? Is there any restriction on which parts/genes the user can choose?
Question4:Does your software let the user create a design by choosing parts/genes from a list/database, such as the Registry? If so, which lists/databases are included? Is there any restriction on which parts/genes the user can choose?
 Answer:Yes. KEGG, partsregistry, Codon usage database, SGD, VFDB, E. coli Genome Project. We have no restriction on genes the user can choose.
Answer:Yes. KEGG, partsregistry, Codon usage database, SGD, VFDB, E. coli Genome Project. We have no restriction on genes the user can choose.
 Question5:Does your software allow users to write new data into any public lists or databases? If so, do you check the new data for errors before allowing it to be written?
Question5:Does your software allow users to write new data into any public lists or databases? If so, do you check the new data for errors before allowing it to be written?
 Answer:No.
Answer:No.
 Question6:Does your software include any other features that encourage the user to create safe designs? Please describe them here.
Question6:Does your software include any other features that encourage the user to create safe designs? Please describe them here.
 Answer:Yes. We develop a plugin to show the relationship between the gene or artificial logic gate gene circuit to guide user to select their target genes safely. Besides, we have reminder about biosafely selection for users.
Answer:Yes. We develop a plugin to show the relationship between the gene or artificial logic gate gene circuit to guide user to select their target genes safely. Besides, we have reminder about biosafely selection for users.
 Question7:Is your team also doing biological work in a wet lab?
Question7:Is your team also doing biological work in a wet lab?
 Answer:No.
Answer:No.
Collaboration
Cooperation: Shenzhen_BGIC_ATCG
We use NucleoMod to assist other wet team, Shenzhen_BGIC_ATCG, to design the biobrick optimized for eukaryons.
Below is one of the examples:
We transform the prokaryon version of cas9 for S.cerevisiae automatically.

Firstly, delete five enzymes of biobrick standard, EcoRI, XbaI, SpeI, PstI, NotI based on synonymous mutation of CDS.
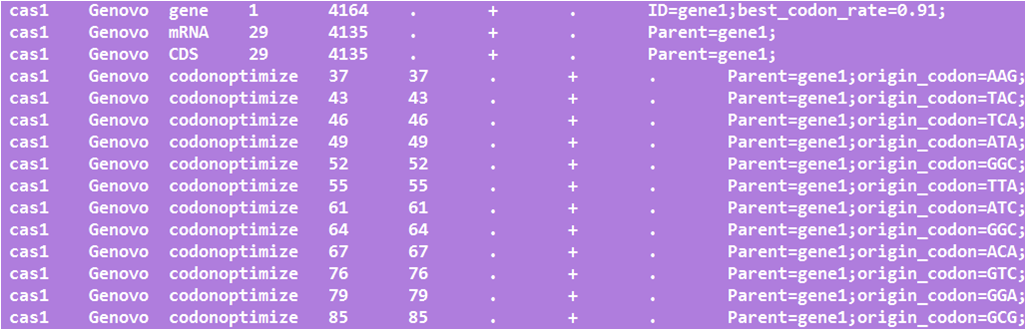
Secondly, optimize the sequence for S.cerevisiae according to codon usage database.
Finally, we recreate the sequence and annotation file.
We tried out SegmMan module to automatically split designed yeast_chr07_4_51 into 2k minichunks in command line.

After run the flow shell, SegmMan analyze the exited enzyme site along the sequence of yeast_chr07_4_51, and finally splited it into about 2k minichunks, so that we can synthesize these fragments and assemble them into whole.
The following is the final result of segmentations. yeast_chr07_4_51 is split into many 2k minichunks. After these fragments are synthesized, they can be assembled into whole step by step, from 2k to 10k to 30k to whole, according Gibson, Goldengate, Telomere Assembly.
On 2k and 10k level, they also have vectors homologous region and designed enzyme site, so that they can be preserved and cut out to use quickly.
 "
"



