Team:Uppsala/killswitches
From 2013.igem.org
| (22 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Team:Uppsala/standard-style | + | {{Team:Uppsala/standard-style-no-frame}} |
| - | + | ||
{{Team:Uppsala/ks-style}} | {{Team:Uppsala/ks-style}} | ||
{{Team:Uppsala/favicon}} | {{Team:Uppsala/favicon}} | ||
| Line 19: | Line 18: | ||
/*Add favicon*/ $("link[rel='shortcut icon']").remove(); $("head").append("<link rel='shortcut icon' type='image/png' href='https://static.igem.org/mediawiki/2013/6/6d/Uppsalas-cow-con.png'>"); }); | /*Add favicon*/ $("link[rel='shortcut icon']").remove(); $("head").append("<link rel='shortcut icon' type='image/png' href='https://static.igem.org/mediawiki/2013/6/6d/Uppsalas-cow-con.png'>"); }); | ||
</script> | </script> | ||
| + | |||
| + | <link href="https://2013.igem.org/Team:Uppsala/lightbox-css-code.css?action=raw&ctype=text/css" type="text/css" rel="stylesheet"> | ||
| + | |||
| + | <script src="https://2013.igem.org/Team:Uppsala/jquery-code.js?action=raw&ctype=text/javascript" type="text/javascript"></script> | ||
| + | |||
| + | <script src="https://2013.igem.org/Team:Uppsala/lightbox-code.js?action=raw&ctype=text/javascript" type="text/javascript"></script> | ||
</head> | </head> | ||
| Line 64: | Line 69: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/metabolic-engineering">Metabolic engineering</a> | <li><a href="https://2013.igem.org/Team:Uppsala/metabolic-engineering">Metabolic engineering</a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/p-coumaric-acid"> | + | <li><a href="https://2013.igem.org/Team:Uppsala/p-coumaric-acid">p-Coumaric acid</a></li> |
<li><a href="https://2013.igem.org/Team:Uppsala/resveratrol">Resveratrol</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/resveratrol">Resveratrol</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/lycopene">Lycopene</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/lycopene">Lycopene</a></li> | ||
| Line 85: | Line 90: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/modeling" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/6/63/Uppsala2013_Modeling.png"></a> | <li><a href="https://2013.igem.org/Team:Uppsala/modeling" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/6/63/Uppsala2013_Modeling.png"></a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/P-Coumaric-acid-pathway"> | + | <li><a href="https://2013.igem.org/Team:Uppsala/P-Coumaric-acid-pathway">Kinetic model</a></li> |
<li><a href="https://2013.igem.org/Team:Uppsala/modeling-tutorial">Modeling tutorial </a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/modeling-tutorial">Modeling tutorial </a></li> | ||
| + | |||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/toxicity-model">Toxicity model</a></li> | ||
</ul></li> | </ul></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/parts" id="list_type2"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/e/eb/Uppsala2013_parts.png"></a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/parts" id="list_type2"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/e/eb/Uppsala2013_parts.png"></a></li> | ||
| Line 107: | Line 114: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/bioart">BioArt</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/bioart">BioArt</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/LactonutritiousWorld">A LactoWorld</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/LactonutritiousWorld">A LactoWorld</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/killswitches">Killswitches</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/realization">Patent</a></li> | ||
</ul></li> | </ul></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a></li> | ||
| Line 124: | Line 133: | ||
<div id="text-box"> | <div id="text-box"> | ||
| - | <p>One major risk | + | <p class="ks">One major risk that the synthetic biology community needs to address is the spread of modified organisms in nature and associated repercusions. Many iGEM teams have addressed this issue by introducing so called killswitches in their design. A killswitch system reduces the risk of bacteria spreading in nature by making it unable to survive without certain conditions. A classical killswitch works by the use of a reagent not present in nature that hinders the expression of a toxin. When the bacteria is released to the enviorment, the reagent is no longer present and the expression of the toxin is released. |
<br><br> | <br><br> | ||
| - | While this sounds like a good solution there are flaws in the system. A large problem is mutations and genetic recombination that destroys genetic code for the toxin, and therefore making the bacteria able to survive once again. We would also like to argue that the modified | + | While this sounds like a good solution there are flaws in the system. A large problem is mutations and genetic recombination that destroys genetic code for the toxin, and therefore making the bacteria able to survive once again. We would also like to argue that the modified bacteria fitness is so much lower than the wild type that the chance of survival in nature is low. To prove this we preformed a competition test, which showed that our modified bacteria would become outcompeted in a couple of days. Therefore we concluded that a killswitch was not needed in our project. |
<br><br> | <br><br> | ||
| - | After a long | + | After a long discussion about killswitches our team reached a couple of conclusions. Our first conclusion is that the best killswitch would be one that removes a function of the bacteria much like <a href="https://2013.igem.org/Team:BGU_Israel/Solution">BGU_Israels p.a.s.e. 2 part</a>. To gain a new function from scratch would need an horizontal genetransfere from another bacteria, or a massive amount of mutations and recombinations. While to loose a function, like producing the toxin, could happen with a single mutation or recombination. Our second conclusion is that the problem with iGEM teams approch to killswitches today is that they do not confirm the effectiveness of the killswitches. How well does the bacteria survive outside the lab? This can easily be tested with an competition test as seen in our project. A killswitch should not simple be inserted as a way to automaticly acheive biosafety criterias without being tested. We hope that future iGEM teams will focus on a low chance of survival outside of the lab, with or without killswitches. |
| - | + | ||
</p></div> | </p></div> | ||
<p> | <p> | ||
| + | <br><br> | ||
We became curious on what other teams thought about killswitches, and decided to convey a survey. With this survey we also wanted to start a debate about killswitches. Can we really trust killswitches completely? What other options are there? Will we ever become 100% safe from spread in nature? | We became curious on what other teams thought about killswitches, and decided to convey a survey. With this survey we also wanted to start a debate about killswitches. Can we really trust killswitches completely? What other options are there? Will we ever become 100% safe from spread in nature? | ||
<br><br> | <br><br> | ||
| Line 146: | Line 155: | ||
<div id="clear"></div> | <div id="clear"></div> | ||
| - | <p>The results where really | + | <p>The results where really fascinating. About a third of the iGEM teams that answered used killswitches, but only half of them belive killswitches will become 100% safe. When asked if we will ever become 100% sure that the bacteria released into nature will be of no danger only one sixth of the teams agreed. The other teams said that you have to weigh the risk versus the benefits. Though there is still not much of a discussion about risks and benefits in iGEM today.</p> |
| - | <p> | + | <p>Our conclusions were that:</p> |
<ul> | <ul> | ||
| - | <li>The main focus of | + | <li>The main focus of biosafety should be to measure the survivabilty of the bacteria in the wild and reducing them if needed.</li> |
| - | <li>The best killswitch is one that | + | <li>The best killswitch is one that eliminate the bacteria when a substrate no longer is added.</li> |
| - | <li>There is a need for a wider discussion about | + | <li>There is a need for a wider discussion about risks and benefits about synthetic biology within iGEM.</li> |
</ul> | </ul> | ||
Latest revision as of 22:37, 28 October 2013
Killswitches
One major risk that the synthetic biology community needs to address is the spread of modified organisms in nature and associated repercusions. Many iGEM teams have addressed this issue by introducing so called killswitches in their design. A killswitch system reduces the risk of bacteria spreading in nature by making it unable to survive without certain conditions. A classical killswitch works by the use of a reagent not present in nature that hinders the expression of a toxin. When the bacteria is released to the enviorment, the reagent is no longer present and the expression of the toxin is released.
While this sounds like a good solution there are flaws in the system. A large problem is mutations and genetic recombination that destroys genetic code for the toxin, and therefore making the bacteria able to survive once again. We would also like to argue that the modified bacteria fitness is so much lower than the wild type that the chance of survival in nature is low. To prove this we preformed a competition test, which showed that our modified bacteria would become outcompeted in a couple of days. Therefore we concluded that a killswitch was not needed in our project.
After a long discussion about killswitches our team reached a couple of conclusions. Our first conclusion is that the best killswitch would be one that removes a function of the bacteria much like BGU_Israels p.a.s.e. 2 part. To gain a new function from scratch would need an horizontal genetransfere from another bacteria, or a massive amount of mutations and recombinations. While to loose a function, like producing the toxin, could happen with a single mutation or recombination. Our second conclusion is that the problem with iGEM teams approch to killswitches today is that they do not confirm the effectiveness of the killswitches. How well does the bacteria survive outside the lab? This can easily be tested with an competition test as seen in our project. A killswitch should not simple be inserted as a way to automaticly acheive biosafety criterias without being tested. We hope that future iGEM teams will focus on a low chance of survival outside of the lab, with or without killswitches.
We became curious on what other teams thought about killswitches, and decided to convey a survey. With this survey we also wanted to start a debate about killswitches. Can we really trust killswitches completely? What other options are there? Will we ever become 100% safe from spread in nature?

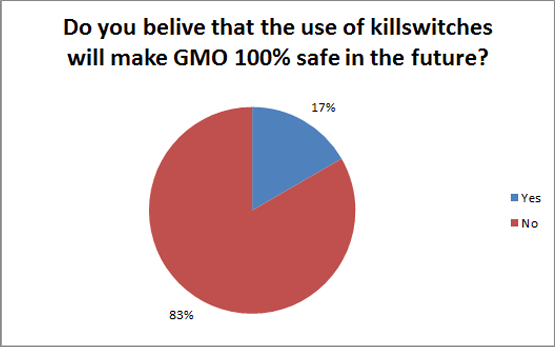
The results where really fascinating. About a third of the iGEM teams that answered used killswitches, but only half of them belive killswitches will become 100% safe. When asked if we will ever become 100% sure that the bacteria released into nature will be of no danger only one sixth of the teams agreed. The other teams said that you have to weigh the risk versus the benefits. Though there is still not much of a discussion about risks and benefits in iGEM today.
Our conclusions were that:
- The main focus of biosafety should be to measure the survivabilty of the bacteria in the wild and reducing them if needed.
- The best killswitch is one that eliminate the bacteria when a substrate no longer is added.
- There is a need for a wider discussion about risks and benefits about synthetic biology within iGEM.
 "
"










