Team:Heidelberg/Templates/Del week10 overview
From 2013.igem.org
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
==Strategy== | ==Strategy== | ||
| - | [[File:Heidelberg_Strategie1.png|300px|right|thumb|Vector map of [[:File:Heidelberg_Psb4k5+BBa J04450 DelRest.gb| pFSN plasmid]] including Primers FS_01 to FS_16 used for assembly of the desired genes from ''D. acidovorans'' genome and the partsregistry backbone pSB4K5 .]]In order to transfer the delftibactin NRPS cluster from ''D. acidovorans'' in ''E. coli'' for efficient production of delftibactin, we designed the plasmid pFSN. It comprises the above listed genes as inserts. Furthermore, it includes an IPTG-inducible promoter, an origin of replication, compatible to those of the plasmids pDelH and pIK8.6 (required for delftibactin expression in ''E. coli'') expression plasmids as well as a kanamycin resistance marker. The choice of the ori was critical, as three plasmids PFSN, pDelH and PIK8.6 would have to be cotransformed in order to obtain the final ''E. coli'' delftibactin production strain and all three plasmids needed to be stabely propagated.<br/>Additionally, mRFP was added as last cds behind the DelRest genes onto the pFSN construct in oder to have an easy control for expression of the DelRest genes located in front of mRFP.<br/><br/> In the Delftibactin cluster the genes DelA, DelB, DelC, DelD, DelE, DelF and DelG are encoded as a single polycistronic operon. However, it is most likely not possible to amplify the entire coding sequence of DelA to DelG in one single PCR amplicon, as its corresponding size of 22.8 kb in combination with the complex GC-rich sequence is most likely not suitable for commonly available polymerases and PCR protocols. Consequently, we will use various primer combinations to amplify the genes in different sub-fragments. These will be assemled again using Gibson cloning<br/><br/>DelO and DelP are encoded in reverse-complementary direction on the Delftibactin cluster. However, in our construct these two genes will be assembled in the same orientation as the other ones and thereby added to the DelA-G operon. As DelO and DelP are also located next to each other and they will therefor be amplified together as a single 2.7 kb amplicon<br/><br/> DelL is the somehow lonesome rider, not surrounded by any of our desired del genes and will thus also be amplified seperately (fragment size 1.4 kb). <br/>As complications are most likely to occure for the very long PCR amplicons, we will start with the huge DelA-G region and optimize PCR conditions for the corresponding amplicons.<br/><br/>We decided to use the DSM-39 substrain of ''Delftia acidovorans'' as our genomic template since our reference paper from Johnston ''et al.'' (2013) showed the gold precipitation expreiments using this substrain. The only full-length genomic sequence available for ''D. acidovorans'' is, however, the SPH-1 substrain sequence. Nevertheless, as the abovementioned paper also based their analysis on the SPH-1 strain sequence, we also based our primer designs on the available SPH-1 substrain genomic sequence, hoping the Del cluster sequence would be highly conserved among different ''D. acidovorans'' strains.< | + | |
| + | [[File:Heidelberg_Strategie1.png|300px|right|thumb|Vector map of [[:File:Heidelberg_Psb4k5+BBa J04450 DelRest.gb| pFSN plasmid]] including Primers FS_01 to FS_16 used for assembly of the desired genes from ''D. acidovorans'' genome and the partsregistry backbone pSB4K5 .]] | ||
| + | |||
| + | <p>In order to transfer the delftibactin NRPS cluster from ''D. acidovorans'' in ''E. coli'' for efficient production of delftibactin, we designed the plasmid pFSN. It comprises the above listed genes as inserts. Furthermore, it includes an IPTG-inducible promoter, an origin of replication, compatible to those of the plasmids pDelH and pIK8.6 (required for delftibactin expression in ''E. coli'') expression plasmids as well as a kanamycin resistance marker. The choice of the ori was critical, as three plasmids PFSN, pDelH and PIK8.6 would have to be cotransformed in order to obtain the final ''E. coli'' delftibactin production strain and all three plasmids needed to be stabely propagated.<br/>Additionally, mRFP was added as last cds behind the DelRest genes onto the pFSN construct in oder to have an easy control for expression of the DelRest genes located in front of mRFP.<br/><br/> In the Delftibactin cluster the genes DelA, DelB, DelC, DelD, DelE, DelF and DelG are encoded as a single polycistronic operon. However, it is most likely not possible to amplify the entire coding sequence of DelA to DelG in one single PCR amplicon, as its corresponding size of 22.8 kb in combination with the complex GC-rich sequence is most likely not suitable for commonly available polymerases and PCR protocols. Consequently, we will use various primer combinations to amplify the genes in different sub-fragments. These will be assemled again using Gibson cloning<br/><br/>DelO and DelP are encoded in reverse-complementary direction on the Delftibactin cluster. However, in our construct these two genes will be assembled in the same orientation as the other ones and thereby added to the DelA-G operon. As DelO and DelP are also located next to each other and they will therefor be amplified together as a single 2.7 kb amplicon<br/><br/> DelL is the somehow lonesome rider, not surrounded by any of our desired del genes and will thus also be amplified seperately (fragment size 1.4 kb). <br/>As complications are most likely to occure for the very long PCR amplicons, we will start with the huge DelA-G region and optimize PCR conditions for the corresponding amplicons.<br/><br/>We decided to use the DSM-39 substrain of ''Delftia acidovorans'' as our genomic template since our reference paper from Johnston ''et al.'' (2013) showed the gold precipitation expreiments using this substrain. The only full-length genomic sequence available for ''D. acidovorans'' is, however, the SPH-1 substrain sequence. Nevertheless, as the abovementioned paper also based their analysis on the SPH-1 strain sequence, we also based our primer designs on the available SPH-1 substrain genomic sequence, hoping the Del cluster sequence would be highly conserved among different ''D. acidovorans'' strains.<br/><br/></p> | ||
| + | |||
| + | ====Plasmid backbone part from the partsregistry==== | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 9: | Line 14: | ||
| pSB4K5 || J04450 || Spring 2012 || 1|| 5G || Backbone for DelA-G,OP,L || Kanamycin | | pSB4K5 || J04450 || Spring 2012 || 1|| 5G || Backbone for DelA-G,OP,L || Kanamycin | ||
|} | |} | ||
| - | <div style="clear:both"></div><br/> | + | <div style="clear:both"></div><br/> |
| + | |||
| + | ====Primer pairs, corresponding sequences and usage==== | ||
| + | |||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| Line 60: | Line 68: | ||
====Results==== | ====Results==== | ||
| - | + | ||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
Latest revision as of 01:28, 29 October 2013
Contents |
Strategy
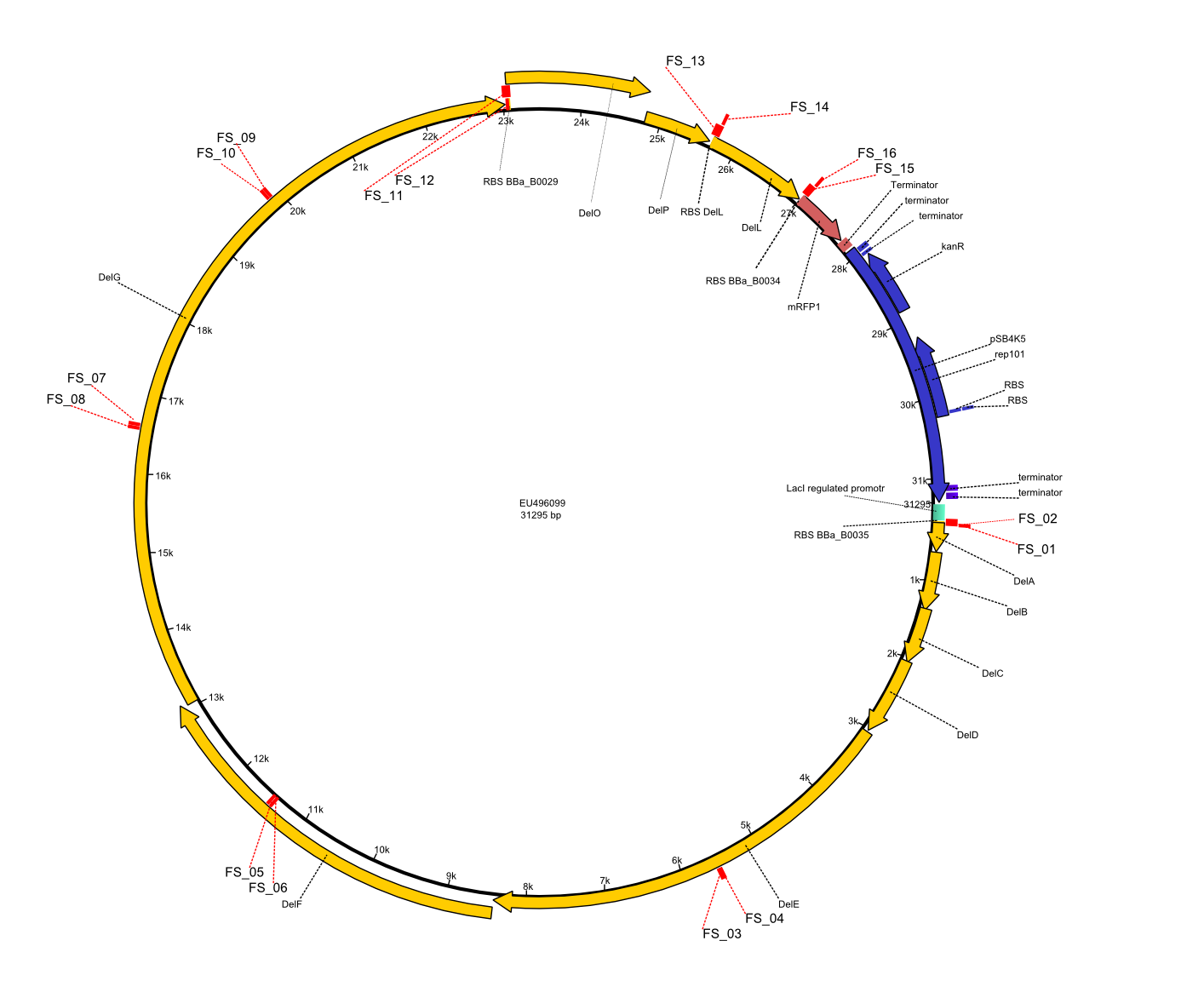
In order to transfer the delftibactin NRPS cluster from D. acidovorans in E. coli for efficient production of delftibactin, we designed the plasmid pFSN. It comprises the above listed genes as inserts. Furthermore, it includes an IPTG-inducible promoter, an origin of replication, compatible to those of the plasmids pDelH and pIK8.6 (required for delftibactin expression in E. coli) expression plasmids as well as a kanamycin resistance marker. The choice of the ori was critical, as three plasmids PFSN, pDelH and PIK8.6 would have to be cotransformed in order to obtain the final E. coli delftibactin production strain and all three plasmids needed to be stabely propagated.
Additionally, mRFP was added as last cds behind the DelRest genes onto the pFSN construct in oder to have an easy control for expression of the DelRest genes located in front of mRFP.
In the Delftibactin cluster the genes DelA, DelB, DelC, DelD, DelE, DelF and DelG are encoded as a single polycistronic operon. However, it is most likely not possible to amplify the entire coding sequence of DelA to DelG in one single PCR amplicon, as its corresponding size of 22.8 kb in combination with the complex GC-rich sequence is most likely not suitable for commonly available polymerases and PCR protocols. Consequently, we will use various primer combinations to amplify the genes in different sub-fragments. These will be assemled again using Gibson cloning
DelO and DelP are encoded in reverse-complementary direction on the Delftibactin cluster. However, in our construct these two genes will be assembled in the same orientation as the other ones and thereby added to the DelA-G operon. As DelO and DelP are also located next to each other and they will therefor be amplified together as a single 2.7 kb amplicon
DelL is the somehow lonesome rider, not surrounded by any of our desired del genes and will thus also be amplified seperately (fragment size 1.4 kb).
As complications are most likely to occure for the very long PCR amplicons, we will start with the huge DelA-G region and optimize PCR conditions for the corresponding amplicons.
We decided to use the DSM-39 substrain of Delftia acidovorans as our genomic template since our reference paper from Johnston et al. (2013) showed the gold precipitation expreiments using this substrain. The only full-length genomic sequence available for D. acidovorans is, however, the SPH-1 substrain sequence. Nevertheless, as the abovementioned paper also based their analysis on the SPH-1 strain sequence, we also based our primer designs on the available SPH-1 substrain genomic sequence, hoping the Del cluster sequence would be highly conserved among different D. acidovorans strains.
Plasmid backbone part from the partsregistry
| Backbone | Part | Distribution | Plate | Well | Usage | Resistance |
|---|---|---|---|---|---|---|
| pSB4K5 | J04450 | Spring 2012 | 1 | 5G | Backbone for DelA-G,OP,L | Kanamycin |
Primer pairs, corresponding sequences and usage
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| FS_01: pSB4K5_DelA_rv | 20-13-06-28 | Amplification of pSB4K5 from the iGEM Distribution. Gibson Primer with overhang to DelA introducing the RBS BBa_B0035 | TCGCGGCGATCCGGTACTGCGCCTCTGTTGAACATCTGATATTCT CCTCTTTAATCGACAGATTGTGTGAAATTGTTATCCGCTCAC |
| FS_02: DelAG_1_fw | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | TTCAACAGAGGCGCAGTACCGGATC |
| FS_03: DelAG_1_rv | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | GTCGGAGACGATGTGGTGCATCAC |
| FS_04: DelAG_2_fw | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | CTGCAGGCCAATGAGCACATCCTG |
| FS_05: DelAG_2_rv | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | CACAGGTGGTAGATGGCGTC |
| FS_06: DelAG_3_fw | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | ATTGCGAGGACTTGCTCGATG |
| FS_07: DelAG_3_rv | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | TTTGCTGCAGCGCCAGCACATCGAG |
| FS_08: DelAG_4_fw | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | GTACGGCCTATCACATCAGCG |
| FS_09: DelAG_4_rv | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | GAAGCTCAGCAGGTTGGGCGAGACG |
| FS_10: DelAG_5_fw | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | GAATTTTGTTCCACCACCTGCTG |
| FS_11: DelAG_5_rv | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. Gibson Primer with overhang to DelOP | CTTGAGCAGGCGCAGTACCTCGGAGGGCGGTCGGCTGGCGTTTTCCATGATTCAGG TTTCCTGTGTGAAGCTCATCTCAGATATCTCCCAGAGTTTCGAGAAAG |
| FS_11: DelAG_5_short_rv | 2013-08-02 | Amplification of DelA-G from Delftia acidovorans genome. | TCAGATATCTCCCAGAGTTTCGAGAAAG |
| FS_12: DelOP_fw | 2013-06-28 | Amplification of DelO-P from Delftia acidovorans genome. | GAATCATGGAAAACGCCAGCCGAC |
| FS_13: DelOP_rv | 2013-06-28 | Amplification of DelO-P from Delftia acidovorans genome. Gibson Primer with overhang to DelL | CAATGTTGGAGGGGCCGAAGCCGATGCCGATCAGCGGGTGGGTTTGCATGGAAGGT CCTTTCATTGGGTCGATGCGTCCAGTGTCACACCGTGGTGTCTGCAGGCG |
| FS_13: DelOP_short_rv | 2013-06-28 | Amplification of DelO-P from Delftia acidovorans genome. Gibson Primer with overhang to DelL | TCACACCGTGGTGTCTGCAGGCG |
| FS_14: DelL_fw | 2013-06-28 | Amplification of DelL from Delftia acidovorans genome. | CAAACCCACCCGCTGATCGGCATC |
| FS_15: DelL_mRFP_pSB4K5_rv | 2013-06-28 | Amplification of DelL from Delftia acidovorans genome. Gibson Primer with overhang to BBa_J04450 | GAAACGCATGAACTCTTTGATAACGTCTTCGGAGGAAGCCATCTAGTATTTCTCCTC TTTCTCTAGTATCAGTCCTGCAGCGCCAGCTGTTCTGTG |
| FS_15: DelL_mRFP_pSB4K5__short_rv | 2013-06-28 | Amplification of DelL from Delftia acidovorans genome. Gibson Primer with overhang to BBa_J04450 | TCAGTCCTGCAGCGCCAGCTGTTCTGTG |
| FS_16: mRFP_pSB4K5_fw (Del) | 2013-06-28 | Amplification of pSB4K5 from iGEM Distribution. Gibson Primer | GCTTCCTCCGAAGACGTTATC |
Amplification of Del Genes from DSM-39 genome
Goals
This week primers FS_01 to FS_16 arrived. As the aim of this week is to amplify the backbone pSB4K5 with Gibson overlaps matching our initial and very last insert fragment, we will firstly use the pSB4K5 backbone including the mRFP cassette as template for this purpose. Furthermore amplification of the desired genes from the Del-cluster of D.acidovorans DSM-39 will be carried out in parallel.
Results
| PCRs from D.acidovorans DSM-39 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelA DelB DelC DelD DelE DelF DelG | DelA-E | FS_02 and FS_03 | ||
| DelA-F | FS_02 and FS_05 | |||
| DelA-G | FS_02 and FS_07 | |||
| DelE-G | FS_04 and FS_07 | |||
| DelF-G | FS_06 and FS_07 | |||
| FS_06 and FS_09 | ||||
| FS_06 and FS_11 | ||||
| DelG | FS_08 and FS_11 | |||
| DelO DelP DelL | DelO-P | FS_12 and FS_13 | ||
| DelL | FS_14 and FS_15 | |||
| pSB4K5 | pSB4K5 | FS_01 and FS_16 | ||
 "
"