Team:Heidelberg/Templates/DelH overview22
From 2013.igem.org
(Difference between revisions)
(Created page with "===Results=== {| class="wikitable" |- ! Colonies !! Alignment File || Sequence |- | III F7 || [[File:Sequencing Result pHM04 VF2 colony DH10b F7.clustal | Sequencing of clone III...") |
|||
| (3 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | |||
===Results=== | ===Results=== | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 4: | Line 5: | ||
! Colonies !! Alignment File || Sequence | ! Colonies !! Alignment File || Sequence | ||
|- | |- | ||
| - | | III F7 || [[File: | + | | III F7 || [[File:Heidelberg_Sequencing Result pHM04 VF2 colony DH10b F7.clustal.txt | Sequencing of clone III F7 with VF2]] || Mutation |
|- | |- | ||
| - | | III F9 || [[File: | + | | III F9 || [[File:Heidelberg_Sequencing Result pHM04 VF2 colony DH10b F9.clustal.txt | Sequencing of clone III F9 with VF2]] || Mutation |
|- | |- | ||
| - | | III G3 || [[File: | + | | III G3 || [[File:Heidelberg_Sequencing Result pHM04 VF2 colony DH10b G3.clustal.txt | Sequencing of clone III G3 with VF2]] || Mutation |
|- | |- | ||
| - | | III G4|| [[File: | + | | III G4|| [[File:Heidelberg_Sequencing Result pHM04 VF2 colony DH10b G4.clustal.txt | Sequencing of clone III G4 with VF2]] || Mutation |
|} | |} | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
<br/> | <br/> | ||
| + | |||
===Vector Maps and Primers=== | ===Vector Maps and Primers=== | ||
| + | |||
'''Strategy A: weak Promotor, weak RBS''' | '''Strategy A: weak Promotor, weak RBS''' | ||
<br/> | <br/> | ||
Expression of the possibly toxic DelH module on pFS02 is minimized by usage of a weak promoter: BBa_J23114 as well as a weak RBS: BBa_0032. | Expression of the possibly toxic DelH module on pFS02 is minimized by usage of a weak promoter: BBa_J23114 as well as a weak RBS: BBa_0032. | ||
| - | [[File: | + | [[File:Heidelberg_pFS_02.png|300px|left|thumb|Vector map of the weak promotor and weak RBS DelH plasmid [[:File:Heidelberg_pFS_02.gb|pFS02 (pSB6A1_BBa_J23114_BBa_B0032_DelH) Gibson plasmid]].]]<div style="clear:both"></div> |
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| Line 49: | Line 52: | ||
|- | |- | ||
| FS_94:BB_HPLC_fw || 26-09-2013 || Primer fw to amplify the backbone pSB6A1, will be used for the ccdB strategy|| ATCTGTTGTTTGTCGGTGAACGC | | FS_94:BB_HPLC_fw || 26-09-2013 || Primer fw to amplify the backbone pSB6A1, will be used for the ccdB strategy|| ATCTGTTGTTTGTCGGTGAACGC | ||
| - | |} | + | |}<div style="clear:both"></div> |
| - | [[File: | + | [[File:Heidelberg_pFS_03.png|300px|left|thumb|Vector map of the DelH plasmid without promoter in pSB4K5 [[:File:Heidelberg_pFS_03.gb|pFS03 (pSB4K5min_KpnI_DelH_BamHI) Gibson plasmid]].]] |
| - | [[File: | + | [[File:Heidelberg_pFS_04.png|300px|none|thumb|Vector map of the pSB6A1 backbone with ccdB cassette [[:File:Heidelberg_pFS_04.gb|pFS04 (pSB6A1_BBa_J23114_BBa_B0032_ccdB_cassette) Gibson plasmid]].]] |
| - | [[File: | + | [[File:Heidelberg_pFS_05.png|300px|none|thumb|Vector map of the final DelH plasmid in pSB6A1 [[:File:Heidelberg_pFS_05.gb|pFS05 (DelH_final_pSB6A1_BBa_J23114_BBa_B0032_KpnI_DelH_BamHI) Gibson plasmid]].]] |
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
<br/> | <br/> | ||
Latest revision as of 02:58, 29 October 2013
Results
| Colonies | Alignment File | Sequence |
|---|---|---|
| III F7 | File:Heidelberg Sequencing Result pHM04 VF2 colony DH10b F7.clustal.txt | Mutation |
| III F9 | File:Heidelberg Sequencing Result pHM04 VF2 colony DH10b F9.clustal.txt | Mutation |
| III G3 | File:Heidelberg Sequencing Result pHM04 VF2 colony DH10b G3.clustal.txt | Mutation |
| III G4 | File:Heidelberg Sequencing Result pHM04 VF2 colony DH10b G4.clustal.txt | Mutation |
Vector Maps and Primers
Strategy A: weak Promotor, weak RBS
Expression of the possibly toxic DelH module on pFS02 is minimized by usage of a weak promoter: BBa_J23114 as well as a weak RBS: BBa_0032.
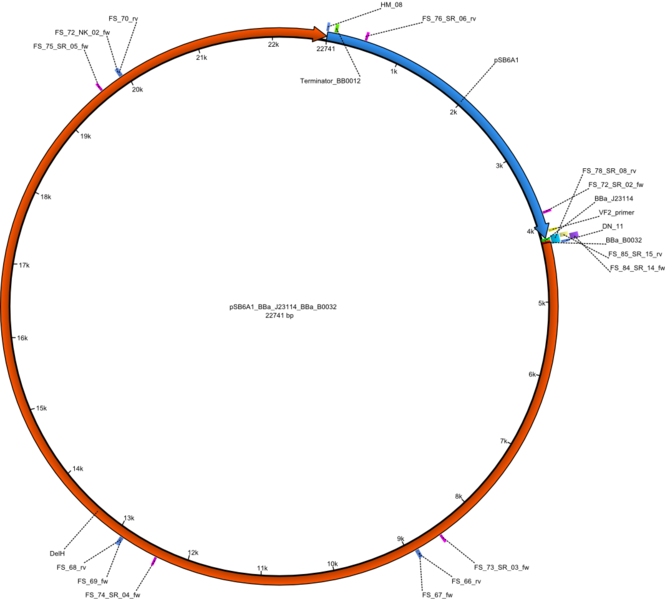
Vector map of the weak promotor and weak RBS DelH plasmid pFS02 (pSB6A1_BBa_J23114_BBa_B0032_DelH) Gibson plasmid.
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| FS_78:BB_HPLC_rev | 26-09-2013 | Gibson-Primer rev, amplify the Backbone pSB6A1 introducing the RBS BBa_B0032 and the promotor BBa_J23114 and creating an overlap to the first fragment of DelH amplified with primer DN_11 | GATTTGGCGCAGGCGGCCACGG TCCATCTAGTACTTTCCTGTGTGACTCTA GAGCTAGCATTGTACCTAGGACTGAGCT AGCCATAAACTCTAGAAGCGGCCGCGAATTC |
| FS_84:BB_HPLC_fw | 26-09-2013 | Gibson-Primer fw to amplify first fragment of DelH introducing the RBS BBa_B0032 and creating an overlap to primer FS_85 thereby partially introducing the promotor BBa_J23114 | GCTCAGTCCTAGGTACAATGCTAGC TCTAGAGTCACACAGGAAAGTACTAGATGGA CCGTGGCCGCCTGCG |
| FS_85:BB_HPLC_rev | 26-09-2013 | Gibson-Primer rev to amplify the Backbone pSB6A1, partially introducing the promotor BBa_B0032 with overlap to primer FS_84 and therefore the promotor BBa_J23114, it creates an overlap to the beginning of DelH | GCGATTTGGCGCAGGCGGCCACGGT CCATCTAGTATTTCTCCTCTTTC |
Strategy B: ccdB Construct
Selection for mutated and thus truncated DelH sequences will be minimized by using a ccdB strategy. DelH will be integrated in a pSB4K5 backbone flanked by KpnI and BamHI sites (pFS02). The pSB6A1 backbone will be assembled with a ccdB cassette flanked by KpnI and BamHI sites (pFS03). The final DelH plasmid will be assembled by restriction - ligation of pFS02 and pFS03 to pFS05.
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| FS_85:BB_HPLC_rev | 26-09-2013 | Gibson-Primer rev to amplify the Backbone pSB6A1, partially introducing the promotor BBa_B0032 with overlap to primer FS_84 and therefore the promotor BBa_J23114, it creates an overlap to the beginning of DelH | GCGATTTGGCGCAGGCGGCCACGGTCCATCTAGTATTTCTCCTCTTTC |
| FS_86:BB_HPLC_rev | 26-09-2013 | Gibson-Primer rev to amplify the Backbone pSB4K5 without any promotor introducing a KpnI cutting site for restriction cloning, creates an overlap to DelH and will be used for the ccdB strategy | GGCGATTTGGCGCAGGCGGCCACGGTCCATGTACTTCGAGTCACTAAGGGCTAAC |
| FS_87:BB_HPLC_fw | 26-09-2013 | Gibson-Primer fw to amplify the Backbone pSB6A1 introducing a BamHI cutting site for restriction cloning and creating an overlap to the last fragment of DelH | CGCTGGAGTACGCGCTGGACTGAGATCCCAGGCATCAAATAAAACG |
| FS_90:ccdB_HPLC_fw | 26-09-2013 | Gibson-Primer fw to amplify the ccdB cassette from the template pDonor Plasmid introducing a KpnI cutting site for restriction cloning, creates an overlap to the promotor BBa_J23114 and will be used for the ccdB strategy | CTCAGTCCTAGGTACAATGCTAGCTCTAGAGTCACACAGGAAAGCAGTAC ACTGGCTGTGTATAAGGGAG |
| FS_93:ccdB_HPLC_rev | 26-09-2013 | Gibson-Primer rev to amplify the ccdB cassette from the template pDonor Plasmid introducing a BamHI cutting site for restriction cloning, creates an overlap to the backbone pSB6A1 and will be used for the ccdB strategy | GTTCACCGACAAACAACAGATGATCCGCGTGGATCCGGCTTAC |
| FS_94:BB_HPLC_fw | 26-09-2013 | Primer fw to amplify the backbone pSB6A1, will be used for the ccdB strategy | ATCTGTTGTTTGTCGGTGAACGC |
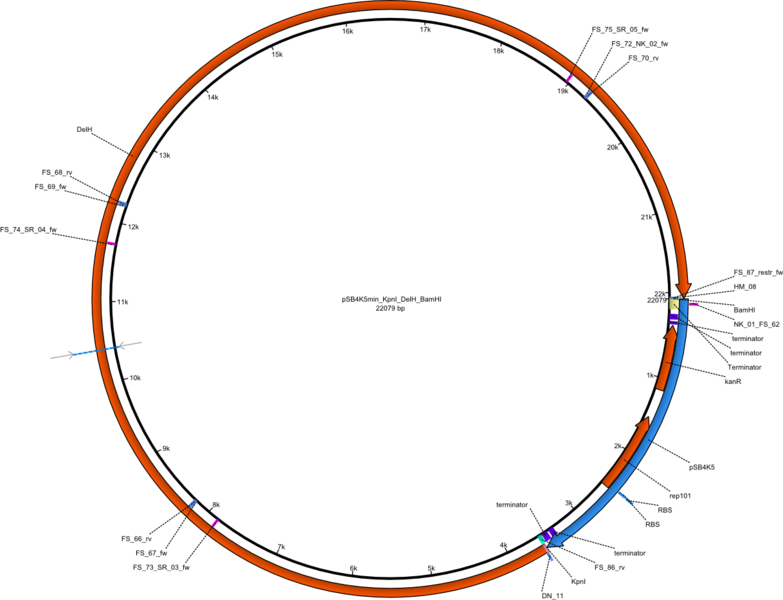
Vector map of the DelH plasmid without promoter in pSB4K5 pFS03 (pSB4K5min_KpnI_DelH_BamHI) Gibson plasmid.
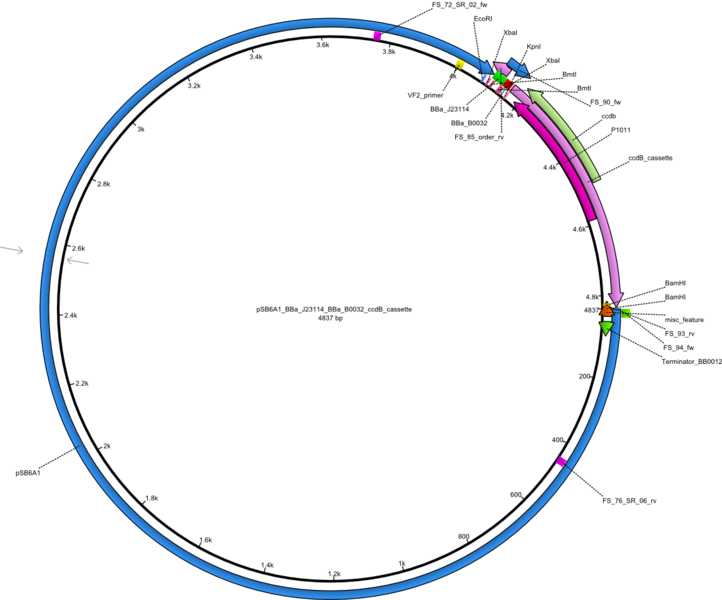
Vector map of the pSB6A1 backbone with ccdB cassette pFS04 (pSB6A1_BBa_J23114_BBa_B0032_ccdB_cassette) Gibson plasmid.
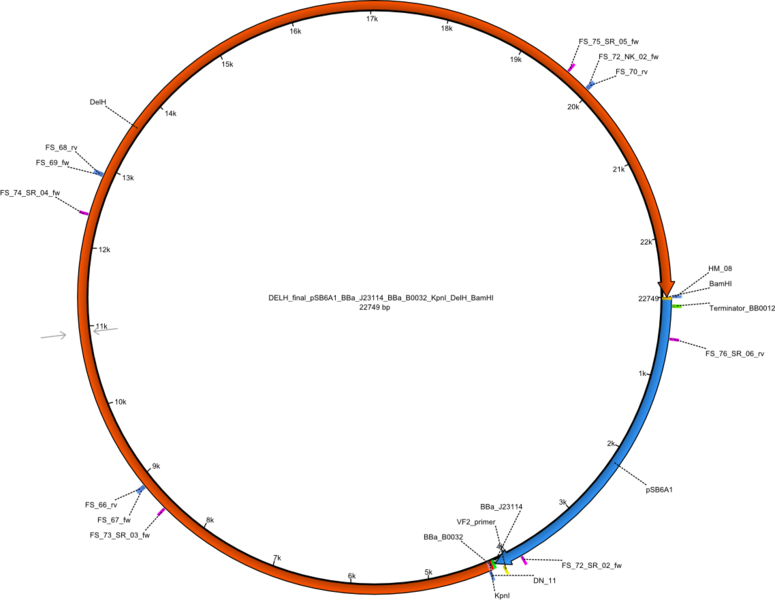
Vector map of the final DelH plasmid in pSB6A1 pFS05 (DelH_final_pSB6A1_BBa_J23114_BBa_B0032_KpnI_DelH_BamHI) Gibson plasmid.
 "
"