Team:Bielefeld-Germany/Labjournal/Molecular
From 2013.igem.org
(Difference between revisions)
| (79 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
__NOTOC__ | __NOTOC__ | ||
| + | |||
<html> | <html> | ||
| - | |||
| - | |||
<style> | <style> | ||
| - | h1{ | + | h1{margin-top:0px;} |
| - | + | ||
| - | + | ||
a{color:rgb(0, 43, 184);} | a{color:rgb(0, 43, 184);} | ||
| - | |||
#leftcol .bigbutton{width:100px; height:40px; line-height:40px; font-size:1.0em; margin-right:10px; display:table;} | #leftcol .bigbutton{width:100px; height:40px; line-height:40px; font-size:1.0em; margin-right:10px; display:table;} | ||
#leftcol .bigbutton a{display:block; height:100%;} | #leftcol .bigbutton a{display:block; height:100%;} | ||
| Line 26: | Line 22: | ||
<h1>Protocols</h1> | <h1>Protocols</h1> | ||
| - | <div id="buttonrow" style="padding-top:30px; padding-bottom:70px; padding-left: | + | <div id="buttonrow" style="padding-top:30px; padding-bottom:70px; padding-left:150px; clear:both;"> |
<div class="bigbutton"> | <div class="bigbutton"> | ||
| - | <p><a href="https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/ | + | <p><a href="https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/ProtocolsPrograms#Genetic Engineering">Genetic Engineering</a></p></div> |
<div class="bigbutton"> | <div class="bigbutton"> | ||
| - | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Labjournal#Analytics">Analytics</a></div> | + | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/ProtocolsPrograms#Analytics">Analytics</a></div> |
<div class="bigbutton"> | <div class="bigbutton"> | ||
| - | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Labjournal# | + | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/ProtocolsPrograms#Cultivation">Cultivations</a></div> |
<div class="bigbutton"> | <div class="bigbutton"> | ||
| - | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Labjournal | + | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/ProtocolsPrograms#Materials">Materials</a></div> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
| Line 50: | Line 43: | ||
==Genetic Engineering== | ==Genetic Engineering== | ||
| + | |||
| + | |||
*In this section of our protocol pages you can read more about our methods for cloning and BioBrick assembly. | *In this section of our protocol pages you can read more about our methods for cloning and BioBrick assembly. | ||
**Genetic engineering is a basic tool of synthetic biology. With the help of standardized DNA building blocks (BioBricks) it is fairly easy to create new and modify existing natural systems. The methods we have used in our project to create BioBricks and to modify, mutate, transform and analyse DNA are presented in this section. | **Genetic engineering is a basic tool of synthetic biology. With the help of standardized DNA building blocks (BioBricks) it is fairly easy to create new and modify existing natural systems. The methods we have used in our project to create BioBricks and to modify, mutate, transform and analyse DNA are presented in this section. | ||
| Line 56: | Line 51: | ||
===Whole Genome Isolation=== | ===Whole Genome Isolation=== | ||
| + | |||
| + | |||
*For the isolation we used a [[Team:Bielefeld-Germany/Labjournal/Molecular#Used Kits | Plasmid Miniprep Kit]]. | *For the isolation we used a [[Team:Bielefeld-Germany/Labjournal/Molecular#Used Kits | Plasmid Miniprep Kit]]. | ||
*Standard Protocol: | *Standard Protocol: | ||
| Line 73: | Line 70: | ||
===Generating electrocompetent cells=== | ===Generating electrocompetent cells=== | ||
| + | |||
| + | |||
*Material: | *Material: | ||
**550 mL LB-Medium | **550 mL LB-Medium | ||
| - | **1 L cooled bidest. | + | **1 L cooled bidest. H<sub>2</sub>O |
**150 mL cooled 10 % glycerine | **150 mL cooled 10 % glycerine | ||
**10 pre-cooled 50 mL Falcons | **10 pre-cooled 50 mL Falcons | ||
| Line 88: | Line 87: | ||
**Divide the cultures into cooled 50 mL Falcons and centrifugate at 4000 rpm, 4 °C for 15 minutes, make sure to slowly accelerate and deccelerate | **Divide the cultures into cooled 50 mL Falcons and centrifugate at 4000 rpm, 4 °C for 15 minutes, make sure to slowly accelerate and deccelerate | ||
**Discard supernatant | **Discard supernatant | ||
| - | **Resuspend pellet in 5 mL cooled bidest | + | **Resuspend pellet in 5 mL cooled bidest H<sub>2</sub>O (and don't get frustrated while doing it, keep shaking gently) |
| - | **Pool two suspensions each, add bidest | + | **Pool two suspensions each, add bidest H<sub>2</sub>O up to 50 mL and centrifugate again (see centrifugation above) |
**Discard supernatant | **Discard supernatant | ||
| - | **Resuspend pellet in 5 mL cooled bidest | + | **Resuspend pellet in 5 mL cooled bidest H<sub>2</sub>O |
| - | **Add bidest | + | **Add bidest H<sub>2</sub>O up to 50 mL and centrifugate again (see centrifugation above) |
**Discard supernatant | **Discard supernatant | ||
**Resuspend pellet in 5 mL cooled 10 % glycerine | **Resuspend pellet in 5 mL cooled 10 % glycerine | ||
| Line 98: | Line 97: | ||
**Discard supernatant | **Discard supernatant | ||
**Add volume of 10 % glycerine that is approximately equal to the volume of the pellet and resuspend | **Add volume of 10 % glycerine that is approximately equal to the volume of the pellet and resuspend | ||
| - | **Divide cells in 50 μL aliquots and freeze in liquid | + | **Divide cells in 50 μL aliquots and freeze in liquid N<sub>2</sub> immediately |
**Store at -80 °C | **Store at -80 °C | ||
===Transformation via electroporation=== | ===Transformation via electroporation=== | ||
| - | *Thaw 50 µL competent E.coli cells on ice, dilute with icecold 50 µL glycerine (10 %) if necessary | + | |
| + | |||
| + | *Thaw 50 µL competent ''E.coli'' cells on ice, dilute with icecold 50 µL glycerine (10 %) if necessary | ||
*Add 0.5-5 µL plasmid to 50 µl electrocompetent cells | *Add 0.5-5 µL plasmid to 50 µl electrocompetent cells | ||
*Store cells on ice for 1 minute | *Store cells on ice for 1 minute | ||
| Line 113: | Line 114: | ||
===Transformation of Single Step (KRX) Competent Cells by Promega=== | ===Transformation of Single Step (KRX) Competent Cells by Promega=== | ||
| + | |||
| + | |||
*Using [http://www.promega.com/~/media/Files/Resources/Protocols/Technical%20Bulletins/101/Single%20Step%20Competent%20Cells%20Protocol.ashx protocol ''E. coli'' KRX single step competent cells by Promega] | *Using [http://www.promega.com/~/media/Files/Resources/Protocols/Technical%20Bulletins/101/Single%20Step%20Competent%20Cells%20Protocol.ashx protocol ''E. coli'' KRX single step competent cells by Promega] | ||
| - | *Remove Single Step (KRX) Competent Cells from –70 °C, and place on ice for 5 minutes or until just thawed | + | *Remove Single Step (KRX) Competent Cells from –70 °C, and place on ice for 5 minutes or until just thawed |
*Add 1–50 ng of DNA (in a volume not greater than 5 μL) to the Single Step (KRX) Competent Cells. Move the pipette tip through the cells while dispensing. Quickly flick the tube several times. Do not vortex! | *Add 1–50 ng of DNA (in a volume not greater than 5 μL) to the Single Step (KRX) Competent Cells. Move the pipette tip through the cells while dispensing. Quickly flick the tube several times. Do not vortex! | ||
*Immediately return the tubes to ice for 5–30 minutes | *Immediately return the tubes to ice for 5–30 minutes | ||
| Line 126: | Line 129: | ||
=== Standard BioBrick Assembly === | === Standard BioBrick Assembly === | ||
| + | |||
| + | |||
*modified from [http://openwetware.org/wiki/Silver:_BB_Strategy Silver lab]: | *modified from [http://openwetware.org/wiki/Silver:_BB_Strategy Silver lab]: | ||
*This assembly method can be used for BioBricks which are bigger than 150 bp. The BioBrick should be at least 500 bp bigger or smaller than the backbone. The BioBrick, which complies with these conditions, is used as the insert and is assembled into the prefix or suffix of the other used BioBrick, called vector. So you have to differentiate between a prefix and a suffix insertion. | *This assembly method can be used for BioBricks which are bigger than 150 bp. The BioBrick should be at least 500 bp bigger or smaller than the backbone. The BioBrick, which complies with these conditions, is used as the insert and is assembled into the prefix or suffix of the other used BioBrick, called vector. So you have to differentiate between a prefix and a suffix insertion. | ||
| + | |||
[[Image:IGEM_Bielefeld_Silver_1.png|300px|thumb|left|Silver Suffix Insertion]] | [[Image:IGEM_Bielefeld_Silver_1.png|300px|thumb|left|Silver Suffix Insertion]] | ||
[[Image:IGEM_Bielefeld_Silver_2.png|300px|thumb|center|Silver Prefix Insertion]] | [[Image:IGEM_Bielefeld_Silver_2.png|300px|thumb|center|Silver Prefix Insertion]] | ||
| + | |||
==== Suffix Insertion ==== | ==== Suffix Insertion ==== | ||
| - | * Digestion of insert: at least 700 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | XbaI]], 1 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | PstI]]. Digest for 1 h at 37 °C, afterwards inactivation for 20 min at 80 °C. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel try to avoid staining or exposure to ultraviolet light of the insert. | + | |
| - | * Digestion of vector about 700 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | SpeI]], 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | PstI]]. Digest for 1 h at 37 °C, afterwards inactivation for 20 min at 80 °C. Add 1 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | AP (Antarctic phosphatase)]] and 1.2 µL 10 x AP reaction buffer, incubate for 1 h at 37 °C. Clean up the vector with a [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_Kits | PCR clean-up kit]]. | + | |
| + | * Digestion of insert: at least 700 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | ''XbaI'']], 1 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | ''PstI'']]. Digest for 1 h at 37 °C, afterwards inactivation for 20 min at 80 °C. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel try to avoid staining or exposure to ultraviolet light of the insert. | ||
| + | * Digestion of vector about 700 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | ''SpeI'']], 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | ''PstI'']]. Digest for 1 h at 37 °C, afterwards inactivation for 20 min at 80 °C. Add 1 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | AP (Antarctic phosphatase)]] and 1.2 µL 10 x AP reaction buffer, incubate for 1 h at 37 °C. Clean up the vector with a [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_Kits | PCR clean-up kit]]. | ||
* Ligation: after digestion and clean-up: 50 - 200 ng of vector, 3 - 10 fold molar access of insert, 20 µL ligation volume, 2 µL T4-Ligase-Buffer, 1 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | T4-Ligase]]. Incubate for 20 - 30 min at room temperature, afterwards inactivation for 5 min at 70 °C. Then: store at -20 °C or transform. | * Ligation: after digestion and clean-up: 50 - 200 ng of vector, 3 - 10 fold molar access of insert, 20 µL ligation volume, 2 µL T4-Ligase-Buffer, 1 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | T4-Ligase]]. Incubate for 20 - 30 min at room temperature, afterwards inactivation for 5 min at 70 °C. Then: store at -20 °C or transform. | ||
==== Prefix Insertion ==== | ==== Prefix Insertion ==== | ||
| - | * Digestion of insert: at least 700 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | EcoRI]], 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | SpeI]]. Digest for 1 h at 37 °C, afterwards inactivation for 20 min at 80 °C. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel try to avoid staining or exposure to ultraviolet light of the insert. | + | |
| - | * Digestion of vector about 700 ng DNA / 10 µL volume, 1 µL 10 x NEBuffer 2.1, 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | EcoRI]], 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | XbaI]]. Digest for 1h at 37 °C, afterwards inactivation for 20 min at 80 °C. Add 1 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | AP (Antarctic phosphatase)]] and 1.2 µL 10 x AP reaction buffer, incubate for 1 h at 37 °C. Clean up the vector with a [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_Kits | PCR clean-up kit]]. | + | |
| + | * Digestion of insert: at least 700 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | ''EcoRI'']], 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | ''SpeI'']]. Digest for 1 h at 37 °C, afterwards inactivation for 20 min at 80 °C. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel try to avoid staining or exposure to ultraviolet light of the insert. | ||
| + | * Digestion of vector about 700 ng DNA / 10 µL volume, 1 µL 10 x NEBuffer 2.1, 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | ''EcoRI'']], 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | ''XbaI'']]. Digest for 1h at 37 °C, afterwards inactivation for 20 min at 80 °C. Add 1 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | AP (Antarctic phosphatase)]] and 1.2 µL 10 x AP reaction buffer, incubate for 1 h at 37 °C. Clean up the vector with a [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_Kits | PCR clean-up kit]]. | ||
* Ligation: after digestion and clean-up: 50 - 200 ng of vector, 3 - 10 fold molar access of insert, 20 µL ligation volume, 2 µL T4-Ligase-Buffer, 1 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | T4-Ligase]]. Incubate for 20 - 30 min at room temperature, afterwards inactivation for 5 min at 70 °C. Then: store at -20 °C or transform. | * Ligation: after digestion and clean-up: 50 - 200 ng of vector, 3 - 10 fold molar access of insert, 20 µL ligation volume, 2 µL T4-Ligase-Buffer, 1 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | T4-Ligase]]. Incubate for 20 - 30 min at room temperature, afterwards inactivation for 5 min at 70 °C. Then: store at -20 °C or transform. | ||
==== Variations ==== | ==== Variations ==== | ||
| + | |||
| + | |||
* A digestion over night is possible. If you digest over night use only 0.1 µL restriction enzyme. | * A digestion over night is possible. If you digest over night use only 0.1 µL restriction enzyme. | ||
* It is also possible to use PCR product as insert. Digest after PCR with corresponding restriction enzymes and clean up with [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_Kits | PCR clean-up kit]]. This could lead to higher yields of insert DNA because a lot of DNA gets lost during the gel electrophoresis clean up. | * It is also possible to use PCR product as insert. Digest after PCR with corresponding restriction enzymes and clean up with [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_Kits | PCR clean-up kit]]. This could lead to higher yields of insert DNA because a lot of DNA gets lost during the gel electrophoresis clean up. | ||
| Line 153: | Line 166: | ||
===NEB Biobrick Assembly Kit=== | ===NEB Biobrick Assembly Kit=== | ||
| - | |||
| - | === Gibson | + | *We used the BioBrick Assembly Kit to assemble an upstream part with a downstream part into destination plasmid. |
| + | |||
| + | |||
| + | [[Image:IGEM_Bielefeld_NEBAssembly_Overview.jpg|300px|thumb|left|NEB BioBrick Assembly Kit.]] | ||
| + | |||
| + | |||
| + | *Digestion Protocoll | ||
| + | **Digest upstream part with [[Team:Bielefeld-Germany/Labjournal/Molecular#Used enzymes|''EcoRI-HF'']] and [[Team:Bielefeld-Germany/Labjournal/Molecular#Used enzymes|''SpeI'']] | ||
| + | **Digest downstream part with [[Team:Bielefeld-Germany/Labjournal/Molecular#Used enzymes|''XbaI'']] and [[Team:Bielefeld-Germany/Labjournal/Molecular#Used enzymes|''PstI'']] | ||
| + | **Digest destiantion destination plasmid with [[Team:Bielefeld-Germany/Labjournal/Molecular#Used enzymes|''EcoRI-HF'']] and [[Team:Bielefeld-Germany/Labjournal/Molecular#Used enzymes|''PstI'']] | ||
| + | ***500 ng part DNA | ||
| + | ***1 μL of each enzyme | ||
| + | ***5 μL 10x NEBuffer 2.1 | ||
| + | ***to 50 μL H<sub>2</sub>0 | ||
| + | **Incubate all three restriction digest reactions at 37°C for 10 minutes and then heat inactivate at 80°C for 20 minutes. | ||
| + | *Dephosphorylation Protocoll | ||
| + | ** Add 1 μL of [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | AP (Antarctic phosphatase)]] and 5 µL of 10x AP reaction buffer to digested destination plasmid, incubate for 1 h at 37 °C. | ||
| + | *Ligation Protocoll | ||
| + | **Ligate the upstream and downstream parts into the digested destination plasmid. | ||
| + | ***2 μL of each part | ||
| + | ***2 μL 10x T4 DNA Ligase Buffer | ||
| + | ***1 μL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used enzymes|T4 DNA Ligase]] | ||
| + | ***11 μL H20 | ||
| + | **Incubate at room temperature for 10 minutes and then heat inactivate at 80°C for 20 minutes. | ||
| + | |||
| + | |||
| + | *[[Team:Bielefeld-Germany/Labjournal/Molecular#Transformation via electroporation|Transform]] 1-2 μL of the ligation product into 50 μL of [[Team:Bielefeld-Germany/Labjournal/Molecular#Generating electrocompetent cells|competent ''E. coli'' cells]]. | ||
| + | |||
| + | |||
| + | === Gibson Assembly === | ||
| + | |||
*Modified from [http://www.nature.com/nmeth/journal/v6/n5/full/nmeth.1318.html Gibson ''et al.'' (2009)] | *Modified from [http://www.nature.com/nmeth/journal/v6/n5/full/nmeth.1318.html Gibson ''et al.'' (2009)] | ||
| + | |||
[[Image:IGEM_Bielefeld_Gibson_Assembly_Principle.jpg|300px|thumb|left|Gibson Assembly]] | [[Image:IGEM_Bielefeld_Gibson_Assembly_Principle.jpg|300px|thumb|left|Gibson Assembly]] | ||
| + | |||
*This assembly method is an isothermal, single-reaction method for assembling multiple overlapping DNA molecules. By coordinating the activity of a 5‘ exonuclease, a DNA polymerase and a DNA ligase two adjacent DNA fragments with complementary terminal sequence overlaps can be joined into a covalently sealed molecule, without the use of any restriction endonuclease. | *This assembly method is an isothermal, single-reaction method for assembling multiple overlapping DNA molecules. By coordinating the activity of a 5‘ exonuclease, a DNA polymerase and a DNA ligase two adjacent DNA fragments with complementary terminal sequence overlaps can be joined into a covalently sealed molecule, without the use of any restriction endonuclease. | ||
| Line 174: | Line 218: | ||
====''In vitro'' recombination==== | ====''In vitro'' recombination==== | ||
| - | * | + | |
| + | |||
| + | * Assembly mixture | ||
*** 320 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#5x isothermal reaction buffer for Gibson assembly| 5x isothermal reaction buffer]] | *** 320 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#5x isothermal reaction buffer for Gibson assembly| 5x isothermal reaction buffer]] | ||
*** 0.64 µL of 10 U mL<sup>-1</sup> [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | T5 exonuclease]] (for DNA molecules overlapping by greater than 150 bp add 3.2 µL of 10 U ml<sup>–1</sup> [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | T5 exonuclease]]) | *** 0.64 µL of 10 U mL<sup>-1</sup> [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | T5 exonuclease]] (for DNA molecules overlapping by greater than 150 bp add 3.2 µL of 10 U ml<sup>–1</sup> [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | T5 exonuclease]]) | ||
| Line 188: | Line 234: | ||
=== Restriction analysis === | === Restriction analysis === | ||
| - | * Digest BioBrick of interest: about 400 ng DNA / 10 µL volume, 1 µL 10x | + | |
| + | |||
| + | * Digest BioBrick of interest: about 400 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL [[Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes | ''NotI'' or ''PstI'']]. Digest for 1 h at 37 °C. ''NotI'' is used to determine the length of the BioBrick and the plasmid backbone, ''PstI'' is used to determine the length of the BioBrick in the plasmid backbone. | ||
* Gel electrophoresis: add 2 µL loading buffer to every digestion mix, apply about 100 - 200 ng DNA / pocket in gel. Don't forget to apply the uncut BioBrick as well. A good agarose concentration for BioBricks between 0.2 and 3 kb is 1.5 %. The smaller your BioBrick of interest is the higher the agarose concentration should be and vice versa. The gel electrophoresis is made with [[Team:Bielefeld-Germany/Labjournal/Molecular#TAE-Buffer | TAE-buffer]]. Be sure that you melt your agarose gel in the same buffer you use for the electrophoresis later. | * Gel electrophoresis: add 2 µL loading buffer to every digestion mix, apply about 100 - 200 ng DNA / pocket in gel. Don't forget to apply the uncut BioBrick as well. A good agarose concentration for BioBricks between 0.2 and 3 kb is 1.5 %. The smaller your BioBrick of interest is the higher the agarose concentration should be and vice versa. The gel electrophoresis is made with [[Team:Bielefeld-Germany/Labjournal/Molecular#TAE-Buffer | TAE-buffer]]. Be sure that you melt your agarose gel in the same buffer you use for the electrophoresis later. | ||
=== Colony PCR === | === Colony PCR === | ||
| + | |||
| + | |||
* Pick one colony with a sterile tip and elute it in 100 µL ddH<sub>2</sub>0 or medium | * Pick one colony with a sterile tip and elute it in 100 µL ddH<sub>2</sub>0 or medium | ||
* Store the colony in 4 °C while colony PCR is running | * Store the colony in 4 °C while colony PCR is running | ||
| Line 216: | Line 266: | ||
| - | + | <br><br> | |
| - | + | ||
==Analytics== | ==Analytics== | ||
| - | *Protocols for the analytical methods we used. | + | |
| - | **DNA and proteins are very small and cannot be seen by the naked eye. To control the success and the results of | + | |
| + | *Protocols for the analytical methods we used are listed in the next chapter. | ||
| + | **DNA and proteins are very small and cannot be seen by the naked eye. To control the success and the results of the upstream and downstream processes, analytical methods are necessary that give reliable results to make DNA or proteins in any way visible for you. The analytical methods we used in our project can be found in this section. | ||
<br><br> | <br><br> | ||
===Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)=== | ===Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)=== | ||
| + | |||
| + | |||
*This analytical method can be used for separation and identification of proteins according to their electrophoretic mobility. The mobility is a function of length of the molecular weight. Proteins that have identical charge per unit mass due to binding of SDS results in an equal electrophoretic mobility. | *This analytical method can be used for separation and identification of proteins according to their electrophoretic mobility. The mobility is a function of length of the molecular weight. Proteins that have identical charge per unit mass due to binding of SDS results in an equal electrophoretic mobility. | ||
====Pouring the polyacrylamide gel==== | ====Pouring the polyacrylamide gel==== | ||
| + | |||
| + | |||
*Make a master mix for the [[Team:Bielefeld-Germany/Labjournal/Molecular#SDS-PAGE gel|stacking]] and [[Team:Bielefeld-Germany/Labjournal/Molecular#SDS-PAGE gel|separating gel]] without adding ammonium persulfate and TEMED. | *Make a master mix for the [[Team:Bielefeld-Germany/Labjournal/Molecular#SDS-PAGE gel|stacking]] and [[Team:Bielefeld-Germany/Labjournal/Molecular#SDS-PAGE gel|separating gel]] without adding ammonium persulfate and TEMED. | ||
*Aliquote 6,5 mL for each separating and 2,5 mL for each stacking gel. | *Aliquote 6,5 mL for each separating and 2,5 mL for each stacking gel. | ||
| - | *Add ammonium persulfate and TEMED to each separating gel aliquote and pour the solution quickly into | + | *Add ammonium persulfate and TEMED to each separating gel aliquote and pour the solution quickly into the gel casting form. Leave about 2 centimeters below the bottom of the comb for the stacking gel. |
*Layer isopropanol on top of the gel. | *Layer isopropanol on top of the gel. | ||
*Leave the separating gel at room temperature for >60 minutes to polymerize. | *Leave the separating gel at room temperature for >60 minutes to polymerize. | ||
*Remove isopropanol and wait until the surface is dry. | *Remove isopropanol and wait until the surface is dry. | ||
| - | *Add ammonium persulfate and TEMED to each | + | *Add ammonium persulfate and TEMED to each stacking gel aliquote and pour the solution quickly into your gel casting form. |
*Insert comb without getting bubbles stuck underneath | *Insert comb without getting bubbles stuck underneath | ||
*Leave the gel at room temperature for >60 minutes to polymerize. | *Leave the gel at room temperature for >60 minutes to polymerize. | ||
| Line 244: | Line 299: | ||
*For storage | *For storage | ||
**Remove sealing and store the gel wrapped in moistened paper towel at 4°C. | **Remove sealing and store the gel wrapped in moistened paper towel at 4°C. | ||
| + | |||
====Preparing the sample==== | ====Preparing the sample==== | ||
| + | |||
| + | |||
*Mix your protein mixture 4:1 with [[Team:Bielefeld-Germany/Labjournal/Molecular#4x Laemmli-buffer| Laemmli-buffer]] (30 mL protein solution + 10 mL Laemmli-buffer) | *Mix your protein mixture 4:1 with [[Team:Bielefeld-Germany/Labjournal/Molecular#4x Laemmli-buffer| Laemmli-buffer]] (30 mL protein solution + 10 mL Laemmli-buffer) | ||
*Heat for 5 minutes at 95 °C. | *Heat for 5 minutes at 95 °C. | ||
| Line 251: | Line 309: | ||
====Running the gel==== | ====Running the gel==== | ||
| + | |||
| + | |||
*Remove sealing, put the polymerized gel into gel box and pour [[Team:Bielefeld-Germany/Labjournal/Molecular#SDS_running_buffer | SDS running buffer]] into the negative and positive electrode chamber. | *Remove sealing, put the polymerized gel into gel box and pour [[Team:Bielefeld-Germany/Labjournal/Molecular#SDS_running_buffer | SDS running buffer]] into the negative and positive electrode chamber. | ||
*Remove comp without destroying the gel pocket. | *Remove comp without destroying the gel pocket. | ||
| - | *Pipet the sample into the gel pockets, adjusting the volume according to the amount of protein in your sample. Make sure to include a lane with molecular weight standards (PageRuler Prestained Protein Ladder™ (Fa. Fermentas)) to determinate the molecular weight of | + | *Pipet the sample into the gel pockets, adjusting the volume according to the amount of protein in your sample. Make sure to include a lane with molecular weight standards (PageRuler Prestained Protein Ladder™ (Fa. Fermentas)) to determinate the molecular weight of the sample. |
*Connect the power lead and run the stacking gel with 10 mA until the blue dye front enters the separating gel. | *Connect the power lead and run the stacking gel with 10 mA until the blue dye front enters the separating gel. | ||
*Raise amperage up to 20 mA for running the separating gel. | *Raise amperage up to 20 mA for running the separating gel. | ||
| Line 260: | Line 320: | ||
===Polyacrylamide gel staining=== | ===Polyacrylamide gel staining=== | ||
| + | |||
| + | |||
====Colloidal Coomassie Blue staining==== | ====Colloidal Coomassie Blue staining==== | ||
| + | |||
| + | |||
Modified staining protocol from [http://http://newjournal.kcsnet.or.kr/main/j_search/j_abstract_view.htm?code=B021105&qpage=j_search&spage=b_bkcs&dpage=ar Kang ''et al''., 2002]. | Modified staining protocol from [http://http://newjournal.kcsnet.or.kr/main/j_search/j_abstract_view.htm?code=B021105&qpage=j_search&spage=b_bkcs&dpage=ar Kang ''et al''., 2002]. | ||
*agitate the [[Team:Bielefeld-Germany/Labjournal/Molecular#Colloidal_Coomassie_Brilliant_Blue_G-250_staining_solution | staining solution]] at 37 °C over night to form the colloids | *agitate the [[Team:Bielefeld-Germany/Labjournal/Molecular#Colloidal_Coomassie_Brilliant_Blue_G-250_staining_solution | staining solution]] at 37 °C over night to form the colloids | ||
| - | *After finishing the SDS-PAGE remove gel from gel casting form and | + | *After finishing the SDS-PAGE remove gel from gel casting form and transfer it in to a box. |
| - | *Add 100 mL of the | + | *Add 100 mL of the staining solution to your polyacrylamid gel. |
*Incubate the gel in the solution at room temperature until the protein bands got an intensive blue color. Shake the gel continuously during incubation. | *Incubate the gel in the solution at room temperature until the protein bands got an intensive blue color. Shake the gel continuously during incubation. | ||
*Remove the staining solution | *Remove the staining solution | ||
| - | * | + | *Wash the gel with dH<sub>2</sub>O |
*Incubate the gel in ddH<sub>2</sub>O (2-6 h) for bleaching the background. Shake the gel continuously during incubation. If necessary replace the colored water with new one. | *Incubate the gel in ddH<sub>2</sub>O (2-6 h) for bleaching the background. Shake the gel continuously during incubation. If necessary replace the colored water with new one. | ||
===MALDI-TOF=== | ===MALDI-TOF=== | ||
| + | |||
| + | |||
*Matrix-assisted Laser Desorption/Ionization – Time of flight (MALDI-TOF) is a procedure to analyze large biomolecules by their mass. | *Matrix-assisted Laser Desorption/Ionization – Time of flight (MALDI-TOF) is a procedure to analyze large biomolecules by their mass. | ||
| + | |||
[[File:IGEM Bielefeld2013 MALDI.png|200px|thumb|left|Our MALDI-TOF device]] | [[File:IGEM Bielefeld2013 MALDI.png|200px|thumb|left|Our MALDI-TOF device]] | ||
| + | |||
| + | |||
*Before measurement, the analyte is co-crystallized on a metal plate within a solid matrix. 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid) and 2,5-dihydroxybenzoic acid (DHB) are most commonly used. | *Before measurement, the analyte is co-crystallized on a metal plate within a solid matrix. 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid) and 2,5-dihydroxybenzoic acid (DHB) are most commonly used. | ||
*Desorption of the analyte is taken by a laser beam, commonly with a wavelength of 337 nm. The matrix absorbs the laser light and the upper layer explosively vaporizes, ejecting both, matrix and analyte molecules. In addition to this vaporization, the laser beam also leads to an ionization of the analytes. The mass analysis is performed by the TOF (time of flight)-analysator. The evaporated ions are accelerated in an electric field. Typical acceleration voltages for this are 10-30 kV. The velocity of an ion depends on its mass and the charge. An ion detector converts the incoming ions into an electrical signal. The advantage of this method is the rapid analysis of a relatively large mass range. | *Desorption of the analyte is taken by a laser beam, commonly with a wavelength of 337 nm. The matrix absorbs the laser light and the upper layer explosively vaporizes, ejecting both, matrix and analyte molecules. In addition to this vaporization, the laser beam also leads to an ionization of the analytes. The mass analysis is performed by the TOF (time of flight)-analysator. The evaporated ions are accelerated in an electric field. Typical acceleration voltages for this are 10-30 kV. The velocity of an ion depends on its mass and the charge. An ion detector converts the incoming ions into an electrical signal. The advantage of this method is the rapid analysis of a relatively large mass range. | ||
| Line 283: | Line 352: | ||
*We used the MALDI-TOF Analysis to characterize the following BioBricks: | *We used the MALDI-TOF Analysis to characterize the following BioBricks: | ||
| - | **< | + | **<bbpart>K1172201</bbpart> |
| - | **< | + | **<bbpart>K1172501</bbpart> |
| + | **<bbpart>K1172303</bbpart> | ||
===Tryptic digest of gel lanes for analysis with MALDI-TOF=== | ===Tryptic digest of gel lanes for analysis with MALDI-TOF=== | ||
| + | |||
| + | |||
*Note: | *Note: | ||
**Make sure to work under a fume hood. | **Make sure to work under a fume hood. | ||
| - | **Do not work with protective gloves to prevent contamination of your sample with | + | **Do not work with protective gloves to prevent contamination of your sample with plasticizers. |
| - | *Reaction tubes have to be cleaned with 60 % (v/v) CH<sub>3</sub>CN | + | *Reaction tubes have to be cleaned with 60 % (v/v) CH<sub>3</sub>CN and 0.1 % (v/v) TFA. Afterwards the solution has to be removed completely followed by evaporation of the tubes under a fume hood. Alternatively microtiter plates from Greiner® (REF 650161) can be used without washing. |
*Cut out the protein lanes of a Coomassie-stained SDS-PAGE using a clean scalpel. Gel parts are transferred to the washed reaction tubes/microtiter plate. If necessary cut the parts to smaller slices. | *Cut out the protein lanes of a Coomassie-stained SDS-PAGE using a clean scalpel. Gel parts are transferred to the washed reaction tubes/microtiter plate. If necessary cut the parts to smaller slices. | ||
*Gel slices should be washed two times. Therefore add 200 µL 30 % (v/v) acetonitrile in 0,1 M ammonium hydrogen carbonate each time and shake lightly for 10 minutes. Remove supernatant and discard to special waste. | *Gel slices should be washed two times. Therefore add 200 µL 30 % (v/v) acetonitrile in 0,1 M ammonium hydrogen carbonate each time and shake lightly for 10 minutes. Remove supernatant and discard to special waste. | ||
| Line 309: | Line 381: | ||
===Preparation and Spotting for analysis of peptides on Bruker AnchorChips=== | ===Preparation and Spotting for analysis of peptides on Bruker AnchorChips=== | ||
| - | *Spot 0,5 - 1 µL sample aliquot | + | |
| + | |||
| + | *Spot 0,5 - 1 µL of sample aliquot | ||
*Add 1 µL HCCA matrix solution to the spotted sample aliquots. Pipet up and down approximately five times to obtain a sufficient mixing. Be careful not to contact the AnchorChip. | *Add 1 µL HCCA matrix solution to the spotted sample aliquots. Pipet up and down approximately five times to obtain a sufficient mixing. Be careful not to contact the AnchorChip. | ||
**Note: Most of the sample solvent needs to be gone in order to achieve a sufficiently low water content. When the matrix solution is added to the previously spotted sample aliquot at a too high water content in the mixture, it will result in undesired crystallization of the matrix outside the anchor spot area. | **Note: Most of the sample solvent needs to be gone in order to achieve a sufficiently low water content. When the matrix solution is added to the previously spotted sample aliquot at a too high water content in the mixture, it will result in undesired crystallization of the matrix outside the anchor spot area. | ||
| Line 315: | Line 389: | ||
*Spot external calibrants on the adjacent calibrant spot positions. Use the calibrant stock solution (Bruker’s “Peptide Calibration Standard II”, Part number #222570), add 125 µL of 0.1% TFA (v/v) in 30% ACN to the vial. Vortex and sonicate the vial. | *Spot external calibrants on the adjacent calibrant spot positions. Use the calibrant stock solution (Bruker’s “Peptide Calibration Standard II”, Part number #222570), add 125 µL of 0.1% TFA (v/v) in 30% ACN to the vial. Vortex and sonicate the vial. | ||
*Mix the calibrant stock solution in a 1:200 ratio with HCCA matrix and deposit 1 µL of the mixture onto the calibrant spots. | *Mix the calibrant stock solution in a 1:200 ratio with HCCA matrix and deposit 1 µL of the mixture onto the calibrant spots. | ||
| + | |||
| + | |||
| + | ===Substrate HPLC=== | ||
| + | |||
| + | *This method has been used for measurement of the substrate concentration | ||
| + | *Protocol: | ||
| + | **Centrifugation (10 min at 14000 g) of the cell culture | ||
| + | **Mix 900 μL of the supernatant with 2 μL 100 g/L sodium azide solution | ||
| + | **Store the sample at - 20 °C or direct measurement with HPLC <br> | ||
| + | *HPLC parameters: | ||
| + | **Stationary phase: 300 mm x 7.8 mm ''NUCLEOGEL SUGAR 810H'' column ([http://www.mn-net.com/HPLCStart/SpecialHPLCphases/Sugars/NUCLEOGELSUGAR810/tabid/10328/language/en-US/Default.aspx ''Macherey Nagel'']) | ||
| + | **Column temperature: 60 °C. | ||
| + | **Eluent: 2.5 mM H<sub>2</sub>SO<sub>4</sub>-solution | ||
| + | **Flow: 0.8 mL min<sup>-1</sup> | ||
| + | **Sample volume: 20 μL with a retention time of 25 min | ||
| + | **Detection: RI detector | ||
| + | **Analysis: Software ''Chromstar 32'' version 6.3 ([http://chromstaranalytical.com/index.htm ''Chromstar Analytical'']) | ||
| + | |||
===NADH-Assay=== | ===NADH-Assay=== | ||
| + | |||
| + | |||
*This method has been used for measurement of intracellular NADH concentration. | *This method has been used for measurement of intracellular NADH concentration. | ||
| Line 326: | Line 420: | ||
**Discard supernatant and wash pellet 3 times with 1 mL of PBS buffer | **Discard supernatant and wash pellet 3 times with 1 mL of PBS buffer | ||
**Resuspend pellet in 1 mL PBS buffer | **Resuspend pellet in 1 mL PBS buffer | ||
| - | **Cell disruption by | + | **Cell disruption by ribolysation (3 x 30 sec at 6500 rpm) |
**Centrifugation for 10 min at maximum speed | **Centrifugation for 10 min at maximum speed | ||
**Store supernatant at - 20 ° C or direct measurement with [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader] | **Store supernatant at - 20 ° C or direct measurement with [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader] | ||
| Line 339: | Line 433: | ||
===Hexadecan Assay=== | ===Hexadecan Assay=== | ||
| + | |||
| + | |||
*This assay has been used to measure cell membrane hydrophobicity. | *This assay has been used to measure cell membrane hydrophobicity. | ||
| Line 354: | Line 450: | ||
| - | ===Preparing samples for Atomic Force Microscopy (AFM) analysis of the | + | ===Preparing samples for Atomic Force Microscopy (AFM) analysis of the outer membrane of cells=== |
| + | |||
| + | |||
| + | *AFM: During the last years, atomic force microscopy has been used increasingly to investigate microbial surfaces at high resolution. The technique provides three-dimensional images of the surface ultrastructure with molecular resolution, in real time, under physiological conditions, and with minimal sample preparation. | ||
| + | **<p align="justify">AFM was carried out using the [http://www.bruker.com/de/products/surface-analysis/atomic-force-microscopy/multimode-8/overview.html MultiMode® 8 AFM from Bruker]. The measurements were performed on air with ‘Tapping Mode’ and in water with ‘Peak Force Mode’.</p> | ||
| + | |||
| + | |||
*Cell preparation: | *Cell preparation: | ||
| - | **Inoculating an overnight culture (30 mL with 1 mL of pre-culture) and | + | **Inoculating an overnight culture (30 mL with 1 mL of pre-culture) and induction of gene expression if necessary at OD<sub> 600</sub> = 1.0. |
| - | **Centrifugation (5 min at 5000 g) of 2 mL overnight culture ( | + | **Centrifugation (5 min at 5000 g) of 2 mL overnight culture (OD<sub> 600</sub> 5 ~ 1 ∙ 10<sup>9</sup> cells/mL). |
**Discard supernatant and wash pellet 3 times with 1 mL of ultra pure water. | **Discard supernatant and wash pellet 3 times with 1 mL of ultra pure water. | ||
| - | **Resuspend pellet in 2 mL ultra pure water (2 ∙ | + | **Resuspend pellet in 2 mL ultra pure water (2 ∙ 10<sup>9</sup> cells). |
| - | *Preparing of AFM glass slides (0.5 | + | *Preparing of AFM glass slides (0.5 cm<sup>2</sup>): |
| Line 370: | Line 472: | ||
**10 minutes sonication of glass slides in ethanol. | **10 minutes sonication of glass slides in ethanol. | ||
**10 minutes sonication of glass slides in ultra pure water. | **10 minutes sonication of glass slides in ultra pure water. | ||
| - | **Dry glass slides under gentle | + | **Dry glass slides under gentle N<sub>2</sub> flow. |
| Line 376: | Line 478: | ||
| - | **Drop 5 μL of a 1:4 dilution of prepared cells (1,25 ∙ | + | **Drop 5 μL of a 1:4 dilution of prepared cells (1,25 ∙ 10<sup>6</sup> cells) to the prepared glass slides for a submonolayer coverage. |
| - | **Dry glass surface with cells under gentle | + | **Dry glass surface with cells under gentle N<sub>2</sub> flow. |
| - | **Wash glass surface three times with ultra pure water and dry between under gentle | + | **Wash glass surface three times with ultra pure water and dry between under gentle N<sub>2</sub> flow. |
**The coated glass slides were kept cool until AFM measurement. | **The coated glass slides were kept cool until AFM measurement. | ||
===NPN membrane permeability assay=== | ===NPN membrane permeability assay=== | ||
| + | |||
| + | |||
*Cell preparation: | *Cell preparation: | ||
| Line 404: | Line 508: | ||
===ONPG membrane permeability assay=== | ===ONPG membrane permeability assay=== | ||
| + | |||
| + | |||
*Cell preparation: | *Cell preparation: | ||
| Line 423: | Line 529: | ||
===Cold osmotic shock=== | ===Cold osmotic shock=== | ||
| + | |||
| + | |||
*Release of periplasmic protein fraction from ''E. coli'' by cold osmotic shock | *Release of periplasmic protein fraction from ''E. coli'' by cold osmotic shock | ||
| Line 441: | Line 549: | ||
===Riboflavin analysis=== | ===Riboflavin analysis=== | ||
| - | = | + | |
| + | |||
| + | '''HPLC-Method''' | ||
| + | |||
| + | * Procedure: | ||
| + | ** Riboflavin was separated from disturbing substances on a C18 column and detected with a fluorescence detector. | ||
| + | * Elution: | ||
| + | ** The sample was injected onto a C18-reversed phase column and eluted with a buffer consisting of 99 % 0.01 M sodium acetate pH 7.4 and 1 % tetrahydrofurane. Total flow is set to 1 ml/min | ||
| + | * Detection: | ||
| + | ** Excitation wavelength of 436 nm and emission wavelength of 535 nm | ||
| + | |||
| + | |||
| + | * C18 reverse phase column | ||
| + | * Isocratic method: 99 % 0.01 M sodium acetate pH 7.4 + 1 % tetrahydrofurane | ||
| + | * Flow = 1 mL min-1 | ||
| + | * UV-detection at 535 nm | ||
| + | * Internal standard: 5.31 x 10^-5 mg L<sup>-1</sup> Riboflavin | ||
| + | * Column: | ||
| + | ** Eurospher II 100-5 C18p by Knauer | ||
| + | ** Dimensions: 150 x 4.6 mm with precolumn | ||
| + | ** Particle size: 5 µm | ||
| + | ** Pore size: 100 Å | ||
| + | ** Material: silica gel | ||
| + | * Software: | ||
| + | ** Clarity (Version 3.0.5.505) by Data Apex | ||
| + | * Autosampler: | ||
| + | ** Midas by Spark Holland | ||
| + | ** Tray cooling: 10 °C | ||
| + | * Pump: | ||
| + | ** L-6200A Intelligent Pump by Hitachi | ||
| + | * UV-Detector: | ||
| + | ** Series 1050 by Hewlett Packard | ||
| + | |||
| + | |||
| + | '''LC-ESI-qTOF-MS(-MS)''' | ||
'''HPLC method''' | '''HPLC method''' | ||
| - | * Column: C18 | + | * Column: C18 reversed phase (Cogent [http://www.mtc-usa.com/diamond_hydride_specs.asp Diamond Hydride]) |
| - | ** dimension: | + | ** dimension: 150 x 2,1 mm |
| - | ** Pore size: | + | ** Pore size: 100 Å |
| - | ** Particle size: | + | ** Particle size: 4 µm |
* Flow: 0.4 mL min<sup>-1</sup> | * Flow: 0.4 mL min<sup>-1</sup> | ||
| - | * Column temperature: | + | * Column temperature: 40 °C |
| - | * | + | * Injection: 5 µl |
| - | ** 0 | + | * Prerun: 5 min |
| - | ** | + | * Eluent: |
| - | ** | + | ** A = 50 % acetonitrile + 50 % H<sub>2</sub>O + 0,1 % formic acid |
| - | ** | + | ** B = 90 % acetonitrile + 10 % H<sub>2</sub>O + 0,1 % formic acid |
| - | ** | + | * Step gradient: |
| + | ** 0:00 min: 0 % eluent A | ||
| + | ** 8:00 min: 100 % eluent A | ||
| + | ** 13:00 min: 100 % eluent A | ||
| + | ** 15:30 min: 0 % eluent A | ||
| + | ** 18:00 min: 0 % eluent A | ||
* VWR Hitachi LaChrom ULTRA HPLC equipment | * VWR Hitachi LaChrom ULTRA HPLC equipment | ||
| - | * Software: HyStar 3.2 | + | * Software: HyStar 3.2, mircrOTOF Control, DataAnalysis 4.0 |
| + | |||
'''Ionization method''' | '''Ionization method''' | ||
| - | * Using Bruker Daltonics micrOTOF<sub>Q</sub> | + | * Using Bruker Daltonics ESI-QTOF micrOTOF<sub>Q</sub> |
| - | * ESI in | + | * ESI-QTOF-MS in positive mode |
* Mass range: 50 - 1500 m/z | * Mass range: 50 - 1500 m/z | ||
| - | * End plate offset: - 500 V, 107 nA | + | *Source: |
| - | * Capillary: | + | ** End plate offset: - 500 V, 107 nA |
| - | * Nebulizer: | + | ** Capillary: - 3500 V, 4 nA |
| - | * Dry gas: 8 L min<sup>-1</sup> | + | ** Nebulizer: 2 bar |
| + | ** Dry gas: 8 L min<sup>-1</sup> | ||
| + | ** Dry Temp.: 180 °C | ||
| + | * Transfer: | ||
| + | ** Funnel 1 RF: 200 Vpp | ||
| + | ** Funnel 2 RF: 300 Vpp | ||
| + | ** ISCID Energy: 0 eV | ||
| + | ** Heyapole RF: 80 Vpp | ||
* Quadrupole | * Quadrupole | ||
| - | ** Ion energy: | + | ** Ion energy: 4 eV |
| - | ** Low mass: | + | ** Low mass: 50 m/z |
| - | * Collision energy: | + | * Collision Cell: |
| - | * Collision RF: | + | ** Collision energy: 8 eV |
| - | * Transfer time: | + | ** Collision RF: 130 Vpp |
| - | * Pre puls storage: | + | ** Transfer time: 75 µs |
| + | ** Pre puls storage: 4 µs | ||
| + | |||
'''MS-MS''' | '''MS-MS''' | ||
| Line 478: | Line 635: | ||
* Collision energy: 30 eV | * Collision energy: 30 eV | ||
| - | |||
| + | ===Redox activity of cytochromes via absorption spectroscopy=== | ||
| + | *Fractioning by [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/Molecular#Cold_osmotic_shock cold osmotic shock] | ||
| + | *Dilution until measured spectrum is within the visible range | ||
| + | *Exposure to air to facilitate oxidation of the cytochromes | ||
| + | *Measurement of oxidized samples | ||
| + | *Addition of a few grains of sodium dithionite | ||
| + | *Wait for approx. 10min | ||
| + | *Measurement of reduced samples | ||
| + | *Comparison of characteristic peaks | ||
| + | *Peak shifts oxidized to reduced | ||
| + | **Soret band: 410nm → 420nm | ||
| + | **β- and α-bands :broad, undefined peak → 525nm and 552nm | ||
| + | *Measurements were performed with the [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader] | ||
| + | |||
| + | <br><br> | ||
==Cultivation== | ==Cultivation== | ||
| + | |||
| + | |||
*These are the protocols for the cultivations and the downstream processing. | *These are the protocols for the cultivations and the downstream processing. | ||
<br><br> | <br><br> | ||
| - | ===Cultivation | + | ===Cultivation Method for UC Davis Collaboration=== |
| - | * | + | *Plasmid-DNA of the Anderson promoters J23101, J23108, J23109 and J23111-J23119 was rehydrated in 10 µL ddH<sub>2</sub>O |
| + | *Plasmid-DNA was transformed into competent cells of ''E.coli''-strain DH5α using transformation protocol | ||
| + | *Each part was cultivated in LB-medium containing 20 mg/L chloramphenicol to an OD<sub>700</sub> of 0.5 at 37°C with 120 rpm orbital shaking | ||
| + | *Strain control was cultivated under the same condutions without chloramphenicol | ||
| + | *Parts were diluted to a starting OD<sub>700</sub> of 0.01 in 200 µL LB and transferred into a clear, flat-bottomed 96-well plate | ||
| + | *Wells were covered by a gas-permeable foil | ||
| + | *The microplate reader(Tecan infinite 200) was ran with the following settings | ||
| + | ** Read duration: 18 hours | ||
| + | ** Read frequency: Once every ten minutes | ||
| + | ** Orbital shaking, amplitude 2.5 mm, frequency 244.5 rpm | ||
| + | ** Absorbance measurement at 700 nm | ||
| + | ** Fluorescence measurement at 588 nm excitation wavelength and 615 nm emission wavelength | ||
| - | |||
| - | |||
| - | |||
| + | ===Cultivation Method for Riboflavin Measurements in supernatans=== | ||
| + | * Most importantly, all cultivations regarding riboflavin production where carried out in [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/ProtocolsPrograms#M9_Medium M9 medium]. | ||
| + | :*One ml of a glycerol culture was poured in 30 ml of M9 medium. Cells were grown overnight. | ||
| + | :*100 ml of M9 medium were inoculated with an adjusted amount of overnight culture, so that the starting cell density was identical in different shaking flasks. | ||
| + | :*Two samples a one ml were transferred to 1,5 ml microcentrifuge tubes. | ||
| + | :*The samples were spun down for 5 min at maximum speed. Afterwards the supernatant was transferred to fresh microcentrifuge tubes. | ||
| + | <br><br> | ||
| + | ==Materials== | ||
| - | |||
| - | |||
*Chemicals, enzymes and kits we used in our lab work. | *Chemicals, enzymes and kits we used in our lab work. | ||
**Chemical and biological reactions need defined conditions to work as expected. The chemicals, enzymes, kits, buffers and media we used in our project are listed in this section. | **Chemical and biological reactions need defined conditions to work as expected. The chemicals, enzymes, kits, buffers and media we used in our project are listed in this section. | ||
| Line 519: | Line 706: | ||
! Enzyme !! style="padding-left:5px;" |Producer | ! Enzyme !! style="padding-left:5px;" |Producer | ||
|- | |- | ||
| - | | DpnI || style="padding-left:5px;" |[http://www.fermentas.de/product_info.php?info=p296 Fermentas] | + | | ''DpnI'' || style="padding-left:5px;" |[http://www.fermentas.de/product_info.php?info=p296 Fermentas] |
|- | |- | ||
| - | | EcoRI || style="padding-left:5px;" |[https://www.neb.com/products/r0101-ecori NEB] | + | | ''EcoRI'' || style="padding-left:5px;" |[https://www.neb.com/products/r0101-ecori NEB] |
|- | |- | ||
| GoTaq DNA-polymerase || style="padding-left:5px;" |[http://www.promega.com/products/pcr/routine-pcr/gotaq-pcr-core-systems/ Promega] | | GoTaq DNA-polymerase || style="padding-left:5px;" |[http://www.promega.com/products/pcr/routine-pcr/gotaq-pcr-core-systems/ Promega] | ||
| Line 529: | Line 716: | ||
| OneTaq DNA-polymerase || style="padding-left:5px;" |[http://www.neb.com/nebecomm/products/productM0480.asp NEB] | | OneTaq DNA-polymerase || style="padding-left:5px;" |[http://www.neb.com/nebecomm/products/productM0480.asp NEB] | ||
|- | |- | ||
| - | | Pfu DNA-polymerase || style="padding-left:5px;" |[http://www.promega.com/products/pcr/routine-pcr/pfu-dna-polymerase/ Promega] | + | | ''Pfu'' DNA-polymerase || style="padding-left:5px;" |[http://www.promega.com/products/pcr/routine-pcr/pfu-dna-polymerase/ Promega] |
|- | |- | ||
| - | | PstI || style="padding-left:5px;" |[https://www.neb.com/products/r0140-psti NEB] | + | | ''PstI'' || style="padding-left:5px;" |[https://www.neb.com/products/r0140-psti NEB] |
|- | |- | ||
| Phusion HF DNA-polymerase || style="padding-left:5px;" |[http://www.finnzymes.com/pcr/phusion_high_fidelity_dna_polymerase.html Finnzymes] | | Phusion HF DNA-polymerase || style="padding-left:5px;" |[http://www.finnzymes.com/pcr/phusion_high_fidelity_dna_polymerase.html Finnzymes] | ||
| Line 537: | Line 724: | ||
| Antarctic phosphatase || style="padding-left:5px;" |[https://www.neb.com/products/m0289-antarctic-phosphatase NEB] | | Antarctic phosphatase || style="padding-left:5px;" |[https://www.neb.com/products/m0289-antarctic-phosphatase NEB] | ||
|- | |- | ||
| - | | SpeI || style="padding-left:5px;" |[https://www.neb.com/products/r0133-spei NEB] | + | | ''SpeI'' || style="padding-left:5px;" |[https://www.neb.com/products/r0133-spei NEB] |
|- | |- | ||
| T4-DNA-Ligase || style="padding-left:5px;" |[https://www.neb.com/products/m0202-t4-dna-ligase NEB] | | T4-DNA-Ligase || style="padding-left:5px;" |[https://www.neb.com/products/m0202-t4-dna-ligase NEB] | ||
| Line 543: | Line 730: | ||
| T5 exonuclease || style="padding-left:5px;" |[http://www.neb.com/nebecomm/products/productM0363.asp NEB] | | T5 exonuclease || style="padding-left:5px;" |[http://www.neb.com/nebecomm/products/productM0363.asp NEB] | ||
|- | |- | ||
| - | | XbaI || style="padding-left:5px;" |[https://www.neb.com/products/r0145-xbai NEB] | + | | ''XbaI'' || style="padding-left:5px;" |[https://www.neb.com/products/r0145-xbai NEB] |
|} | |} | ||
| Line 577: | Line 764: | ||
|- | |- | ||
|} | |} | ||
| - | |||
===Media, buffer, solutions=== | ===Media, buffer, solutions=== | ||
| + | |||
| + | |||
====Chloramphenicol stock solution==== | ====Chloramphenicol stock solution==== | ||
| - | *Solubilize 20 mg mL-1 Chloramphenicol in 100 % Ethanol | + | |
| + | |||
| + | *Solubilize 20 mg mL<sup>-1</sup> Chloramphenicol in 100 % Ethanol | ||
*Store at -20 °C | *Store at -20 °C | ||
| + | |||
====Ampicillin stock solution==== | ====Ampicillin stock solution==== | ||
| + | |||
| + | |||
*Solubilize 100 mg mL<sup>-1</sup> Ampicillin | *Solubilize 100 mg mL<sup>-1</sup> Ampicillin | ||
*Store at -20 °C | *Store at -20 °C | ||
| + | |||
====DNA loading buffer==== | ====DNA loading buffer==== | ||
| + | |||
| + | |||
*50 % (v/v) glycerol | *50 % (v/v) glycerol | ||
*1 mM EDTA | *1 mM EDTA | ||
*0.1 % (w/v) bromphenol blue | *0.1 % (w/v) bromphenol blue | ||
| - | *Solve in | + | *Solve in ddH<sub>2</sub>O |
| + | |||
====LB medium==== | ====LB medium==== | ||
| + | |||
| + | |||
*10 g Trypton | *10 g Trypton | ||
*5 g yeast extract | *5 g yeast extract | ||
| Line 602: | Line 801: | ||
*Adjust pH to 7.4 | *Adjust pH to 7.4 | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | ====SOC medium==== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | *Add the following components for 900 ml of distilled H<sub>2</sub>O: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | *Add the following components for 900 ml of distilled | + | |
**20 g Trypton | **20 g Trypton | ||
**5 g Bacto Yeast Extract | **5 g Bacto Yeast Extract | ||
**2 mL of 5 M NaCl | **2 mL of 5 M NaCl | ||
**2.5 ml of 1 M KCl | **2.5 ml of 1 M KCl | ||
| - | **10 ml of 1 M | + | **10 ml of 1 M MgCl<sub>2</sub> |
**10 ml of 1 M MgSO4 | **10 ml of 1 M MgSO4 | ||
**20 ml of 1 M glucose | **20 ml of 1 M glucose | ||
| - | :Adjust to 1L with distilled | + | :Adjust to 1L with distilled H<sub>2</sub>O. Sterilize by autoclaving. |
====Biotin 500x Stock==== | ====Biotin 500x Stock==== | ||
| + | |||
| + | |||
*Dissolve 20 mg biotin in 100 mL of 0.05 M NaOH solution and filter sterilize | *Dissolve 20 mg biotin in 100 mL of 0.05 M NaOH solution and filter sterilize | ||
*Store at 4 °C | *Store at 4 °C | ||
| Line 678: | Line 825: | ||
====TAE-Buffer==== | ====TAE-Buffer==== | ||
| + | |||
| + | |||
*1 L of 50x TAE buffer | *1 L of 50x TAE buffer | ||
**242.48 g Tris | **242.48 g Tris | ||
| - | **41.02 g | + | **41.02 g sodium acetate |
**18.612 g EDTA | **18.612 g EDTA | ||
**Adjust pH to 7.8 with acetic acid | **Adjust pH to 7.8 with acetic acid | ||
| - | **Solve in | + | **Solve in dH<sub>2</sub>O |
| - | **Dilute 20 mL 50x stock in 1L | + | **Dilute 20 mL 50x stock in 1L dH<sub>2</sub>O for 1x Buffer for PAGE |
====Cell Fractioning Buffers==== | ====Cell Fractioning Buffers==== | ||
| + | |||
=====CFB 1 (pH 8)===== | =====CFB 1 (pH 8)===== | ||
| + | |||
| + | |||
*0.2 M Tris | *0.2 M Tris | ||
| - | *200 g L -1 sucrose | + | *200 g L<sup>-1</sup> sucrose |
*0.1 M EDTA | *0.1 M EDTA | ||
| + | |||
=====CFB 2.1 (pH 8)===== | =====CFB 2.1 (pH 8)===== | ||
| + | |||
| + | |||
*0.01 M Tris | *0.01 M Tris | ||
| - | *0.005 M | + | *0.005 M MgSO<sub>4</sub> |
| + | |||
=====CFB 2.2 (pH 8)===== | =====CFB 2.2 (pH 8)===== | ||
| + | |||
| + | |||
*0.01 M Tris | *0.01 M Tris | ||
| - | *0.005 M | + | *0.005 M MgSO<sub>4</sub> |
*1% Trition | *1% Trition | ||
*2% SDS | *2% SDS | ||
| + | |||
=====CFB 2.3 (pH 8)===== | =====CFB 2.3 (pH 8)===== | ||
| + | |||
| + | |||
*0.01 M Tris | *0.01 M Tris | ||
| - | *0.005 M | + | *0.005 M MgSO<sub>4</sub> |
*1% Trition | *1% Trition | ||
*0.2% SDS | *0.2% SDS | ||
| + | |||
=====CFB 2.4 (pH 8)===== | =====CFB 2.4 (pH 8)===== | ||
| + | |||
| + | |||
*0.01 M Tris | *0.01 M Tris | ||
| - | *0.005 M | + | *0.005 M MgSO<sub>4</sub> |
*1% Trition | *1% Trition | ||
*1% SDS | *1% SDS | ||
| Line 720: | Line 884: | ||
====SDS-PAGE gel==== | ====SDS-PAGE gel==== | ||
| - | The following | + | |
| + | The following amounts are for one gel. | ||
Stacking gel 5 %: | Stacking gel 5 %: | ||
*775 μL H<sub>2</sub>O | *775 μL H<sub>2</sub>O | ||
| Line 728: | Line 893: | ||
*25 μL 10 % Ammonium persulfate | *25 μL 10 % Ammonium persulfate | ||
*3 μL TEMED | *3 μL TEMED | ||
| + | |||
Separating gel 12 %: | Separating gel 12 %: | ||
| Line 739: | Line 905: | ||
====SDS running buffer==== | ====SDS running buffer==== | ||
| + | |||
| + | |||
*25 mM Tris [pH 8,3] | *25 mM Tris [pH 8,3] | ||
*192 mM Glycerol | *192 mM Glycerol | ||
| Line 745: | Line 913: | ||
====4x Laemmli-buffer==== | ====4x Laemmli-buffer==== | ||
| + | |||
| + | |||
*250 mM Tris-HCl | *250 mM Tris-HCl | ||
*40 % [v/v] Glycerol | *40 % [v/v] Glycerol | ||
| Line 753: | Line 923: | ||
====Colloidal Coomassie Brilliant Blue G-250 staining solution==== | ====Colloidal Coomassie Brilliant Blue G-250 staining solution==== | ||
| - | for 1 L staining solution | + | |
| + | |||
| + | *for 1 L staining solution | ||
| + | |||
*dissolve 50 g L<sup>-1</sup> (NH4)<sub>2</sub>SO<sub>4</sub> in ddH<sub>2</sub>O | *dissolve 50 g L<sup>-1</sup> (NH4)<sub>2</sub>SO<sub>4</sub> in ddH<sub>2</sub>O | ||
| Line 763: | Line 936: | ||
====5x isothermal reaction buffer for Gibson assembly==== | ====5x isothermal reaction buffer for Gibson assembly==== | ||
| + | |||
| + | |||
*storage -20˚C | *storage -20˚C | ||
| + | |||
*3 mL of 1 M Tris-HCl (pH 7.5) | *3 mL of 1 M Tris-HCl (pH 7.5) | ||
| - | *150 µL of 2 M | + | *150 µL of 2 M MgCl<sub>2</sub>, |
*60 µL of 100 mM dGTP | *60 µL of 100 mM dGTP | ||
*60 µL of 100 mM dATP | *60 µL of 100 mM dATP | ||
| Line 776: | Line 952: | ||
| + | ====''Geobacter'' Medium==== | ||
| + | *1.5 g NH<sub>4</sub>Cl<sub>2</sub> | ||
| + | *0.6 g Na<sub>2</sub>HPO4 | ||
| + | *0.1 g KCl | ||
| + | *0.82 g Na-acetate | ||
| + | *10 mL 100 X Trace element solution | ||
| + | *10 mL 100 X Vitamin solution | ||
| + | *1 mL 1000 X Selenite-tungstate solution | ||
| + | *8 g Na-fumarate | ||
| + | *2.5 g NaHCO<sub>3</sub> | ||
| + | *980 mL Distilled water | ||
| + | <br> | ||
| + | *'''100 X Trace element solution''' | ||
| + | **1.5 g Nitrilotriacetic acid | ||
| + | **3 g MgSO<sub>4</sub> x 7 H<sub>2</sub>O | ||
| + | **0.5 g MnSO<sub>4</sub> x H<sub>2</sub>O | ||
| + | **1 g NaCl | ||
| + | **0.1 g FeSO<sub>4</sub> x 7 H<sub>2</sub>O | ||
| + | **0.18 g CoSO<sub>4</sub> x 7 H<sub>2</sub>O | ||
| + | **0.1 g CaCl<sub>2</sub> x 2 H<sub>2</sub>O | ||
| + | **0.18 g ZnSO<sub>4</sub> x 7 H<sub>2</sub>O | ||
| + | **0.01 g CuSO<sub>4</sub> x 5 H<sub>2</sub>O | ||
| + | **0.02 g KAl(SO<sub>4</sub>)<sub>2</sub> x 12 H<sub>2</sub>O | ||
| + | **0.01 g H<sub>3</sub>BO<sub>3</sub> | ||
| + | **0.01 g Na<sub>2</sub>MoO4 x 2 H<sub>2</sub>O | ||
| + | **0.03 g NiCl<sub>2</sub> x 2 H<sub>2</sub>O | ||
| + | **0.3 mg Na<sub>2</sub>SeO<sub>3</sub> | ||
| + | **1000 mL Distilled water | ||
| + | <br> | ||
| + | :*First dissolve nitrilotriacetic acid and adjust pH to 6.5 with KOH, then add minerals and adjust final pH to 7.0 with KOH. | ||
| + | <br> | ||
| + | *'''100 X Vitamin Solution''' | ||
| + | **2 mg Biotin | ||
| + | **2 mg Folic acid | ||
| + | **10 mg Pyridoxine-HCl | ||
| + | **5 mg Thiamine-HCl x 2 H<sub>2</sub>O | ||
| + | **5 mg Riboflavin | ||
| + | **5 mg Nicotinic acid | ||
| + | **5 mg D-Ca-pantothenate | ||
| + | **0,1 mg Vitamin B<sub>12</sub> | ||
| + | **5 mg p-Aminobenzoic acid | ||
| + | **5 mg Lipoic acid<sub>12</sub> | ||
| + | **1000 mL Distilled water | ||
| + | <br> | ||
| + | *'''1000 X Selenite-tungstate solution''' | ||
| + | **0,5 g NaOH | ||
| + | **3 mg Na<sub>2</sub>SeO3 x 5 H<sub>2</sub>O | ||
| + | **4 mg Na<sub>2</sub>WO4 x H<sub>2</sub>O | ||
| + | **1000 mL Distilled water | ||
| + | <br> | ||
| + | :*Dissolve all ingredients, except fumarate and NaHCO<sub>3</sub>, bring to a boil, cool down to room temperature while gassing the medium with N<sub>2</sub> and add the NaHCO<sub>3</sub>. Add filter sterilized medium and 0.5 mL/10mL medium from an anoxic filter sterilized 16 % w/v Na-fumarate stock solution to autoclaved cultivation-tubes by injecting it through the silicone stoppers, using a second cannula to prevent overpressure. | ||
| + | <br> | ||
| + | |||
| + | ====M9 Medium==== | ||
| + | *To prepare 1 liter of M9 minimal medium add the following components to 836.7 mL sterile distilled water. To avoid precipitation, start with CaCl<sub>2</sub> and mix the solution thoroughly after addition of a component. | ||
| + | <br> | ||
| + | *100 mL 10 X M9 salt solution | ||
| + | *50 mL 20 X carbon source | ||
| + | *10 mL 100 X trace element solution | ||
| + | *1 mL autoclaved 1 M MgSO<sub>4</sub> | ||
| + | *0.3 mL autoclaved 1 M CaCl<sub>2</sub> | ||
| + | *1 mL filter sterilized 1 g/L biotin | ||
| + | *1 mL filter sterilized 1 g/L thiamin | ||
| + | <br> | ||
| + | |||
| + | *'''10X M9 salt stock solution''' | ||
| + | **75.2 g Na<sub>2</sub>HPO<sub>4</sub> x 2H<sub>2</sub>O | ||
| + | **30 g KH<sub>2</sub>PO<sub>4</sub> | ||
| + | **5 g NaCl | ||
| + | **5 g NH<sub>4</sub>Cl | ||
| + | **Dissolve the salts in 800 mL water and adjust the pH to 7.2 with NaOH. Add water to a final volume of 1 L and autoclave for 15 minutes at 121 °C. | ||
| + | <br> | ||
| + | *'''20X Carbon source stock solution''' | ||
| + | |||
| + | <div align="center"> | ||
| + | {| class="wikitable" style="text-align:center" | ||
| + | |- | ||
| + | !carbon source<br> - !! Concentration<br> g/L !! Concentration<br> mol/L !! Concentration<br> mol<sub>C</sub>/L !! Final concentration<br> mmol/L | ||
| + | |- | ||
| + | |glucose || 200 || 1,11||rowspan="3"| 6,66|| 55,55 | ||
| + | |- | ||
| + | |glycerol ||204,46|| 2,22 || 111 | ||
| + | |- | ||
| + | |NA-acetate ||273,15 ||3,33 || 166,5 | ||
| + | |} | ||
</div> | </div> | ||
| + | <br> | ||
| + | :*To produce a 20X stock solution for a single carbon source dissolve the corresponding amount of the carbon source in distilled water and autoclave for 15 minutes at 121 °C. | ||
| + | <br> | ||
| + | *'''100X trace element solution''' | ||
| + | **5 g EDTA | ||
| + | **0,83 g FeCl<sub>3</sub> x 6 H<sub>2</sub>O | ||
| + | **84 mg ZnCl<sub>2</sub> | ||
| + | **13 mg CuCl<sub>2</sub> x 2 H<sub>2</sub>O | ||
| + | **10 mg CoCl<sub>2</sub> x 6 H<sub>2</sub>O | ||
| + | **10 mg H<sub>3</sub>BO<sub>3</sub> | ||
| + | **1,6 mg MnCl<sub>2</sub> x 4 H<sub>2</sub>O | ||
| + | **Dissolve 5g EDTA in 800 ml water and adjust the pH to 7.5 with NaOH. Then add the other components and add distilled water to a final volume of 1 L. | ||
| + | **Sterilize the solution over a 0.22-μm filter | ||
| - | <div id=" | + | <br><br><br><br> |
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="withMenu" id="asdf"> | ||
| + | <html> | ||
| + | <div id="nav2" style="width:210px; padding-bottom:5px; padding-left:15px;"> | ||
| + | |||
| + | <div class="navbutton" id="home" style="float:left; padding-left:55px; margin-left:10px; padding-top:0px;"> | ||
| + | <a href="https://2013.igem.org/Team:Bielefeld-Germany" title="Jump to Frontpage"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/f/f6/Bielefeld-Germany2013-ButtonHome.png" height="15px"> | ||
| + | </a> | ||
| + | </div> | ||
| + | <div class="navbutton" id="top" style="float:left; padding-left:10px; margin-left:10px; padding-top:0px;"> | ||
| + | <a href="#" title="Jump to top"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/a/ab/Bielefeld-Germany2013-Up_orange_new.png" height="15px"> | ||
| + | </a> | ||
| + | </div> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | <div id="rightcol" style="width:210px; height:100%; overflow-y:auto; box-shadow:0px 0px 2px 0px grey;" padding:0px 20px;> | ||
__TOC__ | __TOC__ | ||
| - | + | </div> | |
| + | |||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 03:26, 29 October 2013
Protocols
Genetic Engineering
- In this section of our protocol pages you can read more about our methods for cloning and BioBrick assembly.
- Genetic engineering is a basic tool of synthetic biology. With the help of standardized DNA building blocks (BioBricks) it is fairly easy to create new and modify existing natural systems. The methods we have used in our project to create BioBricks and to modify, mutate, transform and analyse DNA are presented in this section.
Whole Genome Isolation
- For the isolation we used a Plasmid Miniprep Kit.
- Standard Protocol:
- Centrifuge 10 mL of over-night liquid culture
- Resuspend pellet in 800 µL of resuspension solution
- Ribolyse three times for 60 s with 6500 rpm (30 s break between every run, 400 mg beads)
- Centrifuge 3 min at 10,000 rpm
- Transfer 500 µL of supernatant into new 2 mL tube
- Add 500 µL of lysis buffer, invert 4 - 6 times
- Add 700 µL of neutralisation buffer, invert 4 - 6 times
- Centrifuge 10 min at 12,000 rpm
- Transfer 2 x 600 µL of supernatant into spin column (centrifuge down each time for 30 s at 12,000 rpm)
- Two wash steps with 750 µL of washing buffer
- Dry column
- Elute in 75 µL of elution buffer
Generating electrocompetent cells
- Material:
- 550 mL LB-Medium
- 1 L cooled bidest. H2O
- 150 mL cooled 10 % glycerine
- 10 pre-cooled 50 mL Falcons
- Protocol:
- Inoculate 2x3 mL LB with bacterial stock; incubate over night at 37 °C and 200 rpm
- Inoculate 2x250 mL LB with the over night cultures in 1-litre-flask at 37 °C and 140 rpm
- Incubate until OD600 0,4-0,6
- Cool the culture 15-30 minutes on ice
- Onwards all steps at 4°C
- Divide the cultures into cooled 50 mL Falcons and centrifugate at 4000 rpm, 4 °C for 15 minutes, make sure to slowly accelerate and deccelerate
- Discard supernatant
- Resuspend pellet in 5 mL cooled bidest H2O (and don't get frustrated while doing it, keep shaking gently)
- Pool two suspensions each, add bidest H2O up to 50 mL and centrifugate again (see centrifugation above)
- Discard supernatant
- Resuspend pellet in 5 mL cooled bidest H2O
- Add bidest H2O up to 50 mL and centrifugate again (see centrifugation above)
- Discard supernatant
- Resuspend pellet in 5 mL cooled 10 % glycerine
- Transfer suspensions in two 50 mL Falcons and centrifugate again (see centrifugation above)
- Discard supernatant
- Add volume of 10 % glycerine that is approximately equal to the volume of the pellet and resuspend
- Divide cells in 50 μL aliquots and freeze in liquid N2 immediately
- Store at -80 °C
Transformation via electroporation
- Thaw 50 µL competent E.coli cells on ice, dilute with icecold 50 µL glycerine (10 %) if necessary
- Add 0.5-5 µL plasmid to 50 µl electrocompetent cells
- Store cells on ice for 1 minute
- Electroporate at U = 2.5 kV, C = 25 µF, R = 400 Ώ
- Transfer transformation reaction to 450 µL SOC-Medium and shake 1 h at 37 °C
- Centrifuge 2 min at 2000 rpm and plate on selective LB-Medium
- Incubate over night at 37 °C
Transformation of Single Step (KRX) Competent Cells by Promega
- Using [http://www.promega.com/~/media/Files/Resources/Protocols/Technical%20Bulletins/101/Single%20Step%20Competent%20Cells%20Protocol.ashx protocol E. coli KRX single step competent cells by Promega]
- Remove Single Step (KRX) Competent Cells from –70 °C, and place on ice for 5 minutes or until just thawed
- Add 1–50 ng of DNA (in a volume not greater than 5 μL) to the Single Step (KRX) Competent Cells. Move the pipette tip through the cells while dispensing. Quickly flick the tube several times. Do not vortex!
- Immediately return the tubes to ice for 5–30 minutes
- Heat-shock cells for 15–20 seconds in a water bath at exactly 42 °C. Do not shake.
- Immediately place the tubes on ice for 2 minutes.
- Add 450 μL of room-temperature SOC-Medium to each transformation reaction, and incubate for 60 minutes at 37 °C with shaking (approximately 225 rpm). For best transformation efficiency, lay the tubes on their sides and tape them to the platform.
- For each transformation reaction, we recommend plating 100 μL of undiluted cells and 1:10 and 1:100 cell dilutions on antibiotic plates. Incubate the plates at 37 °C over night.
Standard BioBrick Assembly
- modified from [http://openwetware.org/wiki/Silver:_BB_Strategy Silver lab]:
- This assembly method can be used for BioBricks which are bigger than 150 bp. The BioBrick should be at least 500 bp bigger or smaller than the backbone. The BioBrick, which complies with these conditions, is used as the insert and is assembled into the prefix or suffix of the other used BioBrick, called vector. So you have to differentiate between a prefix and a suffix insertion.
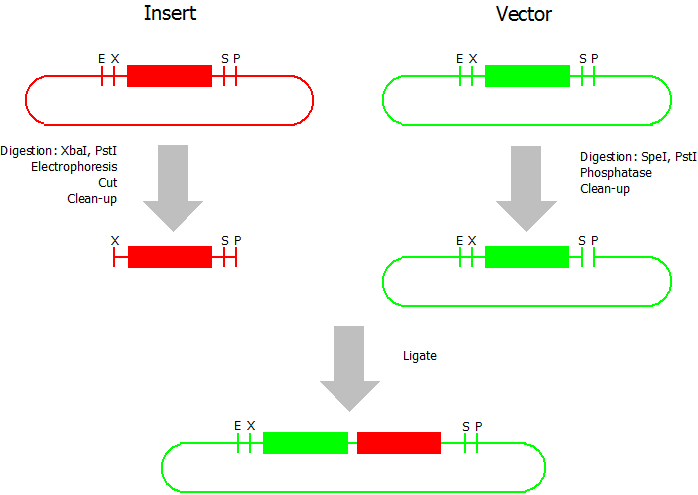
Suffix Insertion
- Digestion of insert: at least 700 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL XbaI, 1 µL PstI. Digest for 1 h at 37 °C, afterwards inactivation for 20 min at 80 °C. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel try to avoid staining or exposure to ultraviolet light of the insert.
- Digestion of vector about 700 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL SpeI, 0.5 µL PstI. Digest for 1 h at 37 °C, afterwards inactivation for 20 min at 80 °C. Add 1 µL AP (Antarctic phosphatase) and 1.2 µL 10 x AP reaction buffer, incubate for 1 h at 37 °C. Clean up the vector with a PCR clean-up kit.
- Ligation: after digestion and clean-up: 50 - 200 ng of vector, 3 - 10 fold molar access of insert, 20 µL ligation volume, 2 µL T4-Ligase-Buffer, 1 µL T4-Ligase. Incubate for 20 - 30 min at room temperature, afterwards inactivation for 5 min at 70 °C. Then: store at -20 °C or transform.
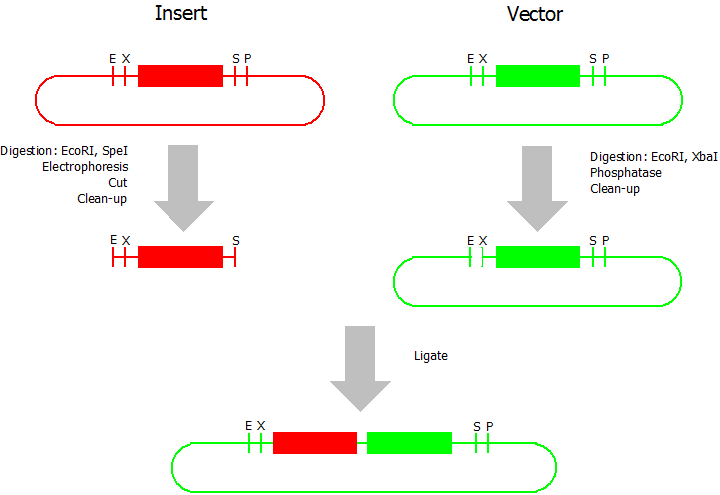
Prefix Insertion
- Digestion of insert: at least 700 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL EcoRI, 0.5 µL SpeI. Digest for 1 h at 37 °C, afterwards inactivation for 20 min at 80 °C. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel try to avoid staining or exposure to ultraviolet light of the insert.
- Digestion of vector about 700 ng DNA / 10 µL volume, 1 µL 10 x NEBuffer 2.1, 0.5 µL EcoRI, 0.5 µL XbaI. Digest for 1h at 37 °C, afterwards inactivation for 20 min at 80 °C. Add 1 µL AP (Antarctic phosphatase) and 1.2 µL 10 x AP reaction buffer, incubate for 1 h at 37 °C. Clean up the vector with a PCR clean-up kit.
- Ligation: after digestion and clean-up: 50 - 200 ng of vector, 3 - 10 fold molar access of insert, 20 µL ligation volume, 2 µL T4-Ligase-Buffer, 1 µL T4-Ligase. Incubate for 20 - 30 min at room temperature, afterwards inactivation for 5 min at 70 °C. Then: store at -20 °C or transform.
Variations
- A digestion over night is possible. If you digest over night use only 0.1 µL restriction enzyme.
- It is also possible to use PCR product as insert. Digest after PCR with corresponding restriction enzymes and clean up with PCR clean-up kit. This could lead to higher yields of insert DNA because a lot of DNA gets lost during the gel electrophoresis clean up.
- Sometimes some BioBricks are hard to assemble. Then you have to clean up the vector by gel electrophoresis as well.
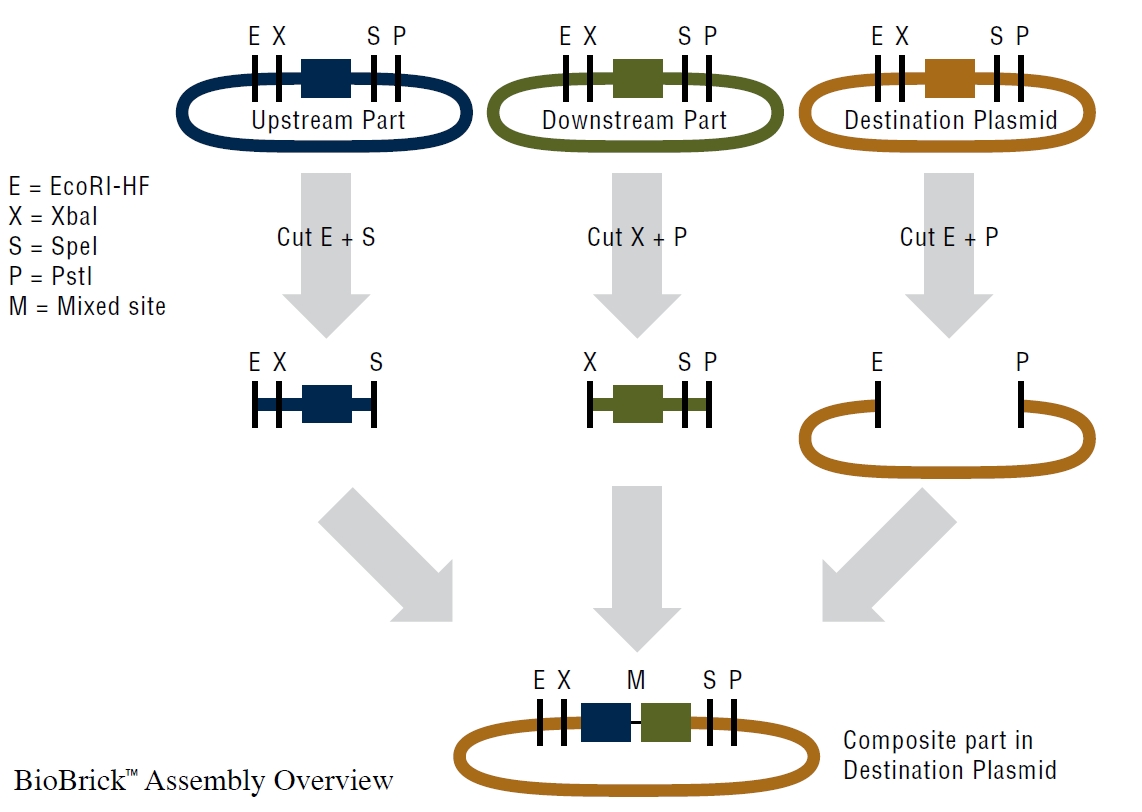
NEB Biobrick Assembly Kit
- We used the BioBrick Assembly Kit to assemble an upstream part with a downstream part into destination plasmid.
- Digestion Protocoll
- Digest upstream part with EcoRI-HF and SpeI
- Digest downstream part with XbaI and PstI
- Digest destiantion destination plasmid with EcoRI-HF and PstI
- 500 ng part DNA
- 1 μL of each enzyme
- 5 μL 10x NEBuffer 2.1
- to 50 μL H20
- Incubate all three restriction digest reactions at 37°C for 10 minutes and then heat inactivate at 80°C for 20 minutes.
- Dephosphorylation Protocoll
- Add 1 μL of AP (Antarctic phosphatase) and 5 µL of 10x AP reaction buffer to digested destination plasmid, incubate for 1 h at 37 °C.
- Ligation Protocoll
- Ligate the upstream and downstream parts into the digested destination plasmid.
- 2 μL of each part
- 2 μL 10x T4 DNA Ligase Buffer
- 1 μL T4 DNA Ligase
- 11 μL H20
- Incubate at room temperature for 10 minutes and then heat inactivate at 80°C for 20 minutes.
- Ligate the upstream and downstream parts into the digested destination plasmid.
- Transform 1-2 μL of the ligation product into 50 μL of competent E. coli cells.
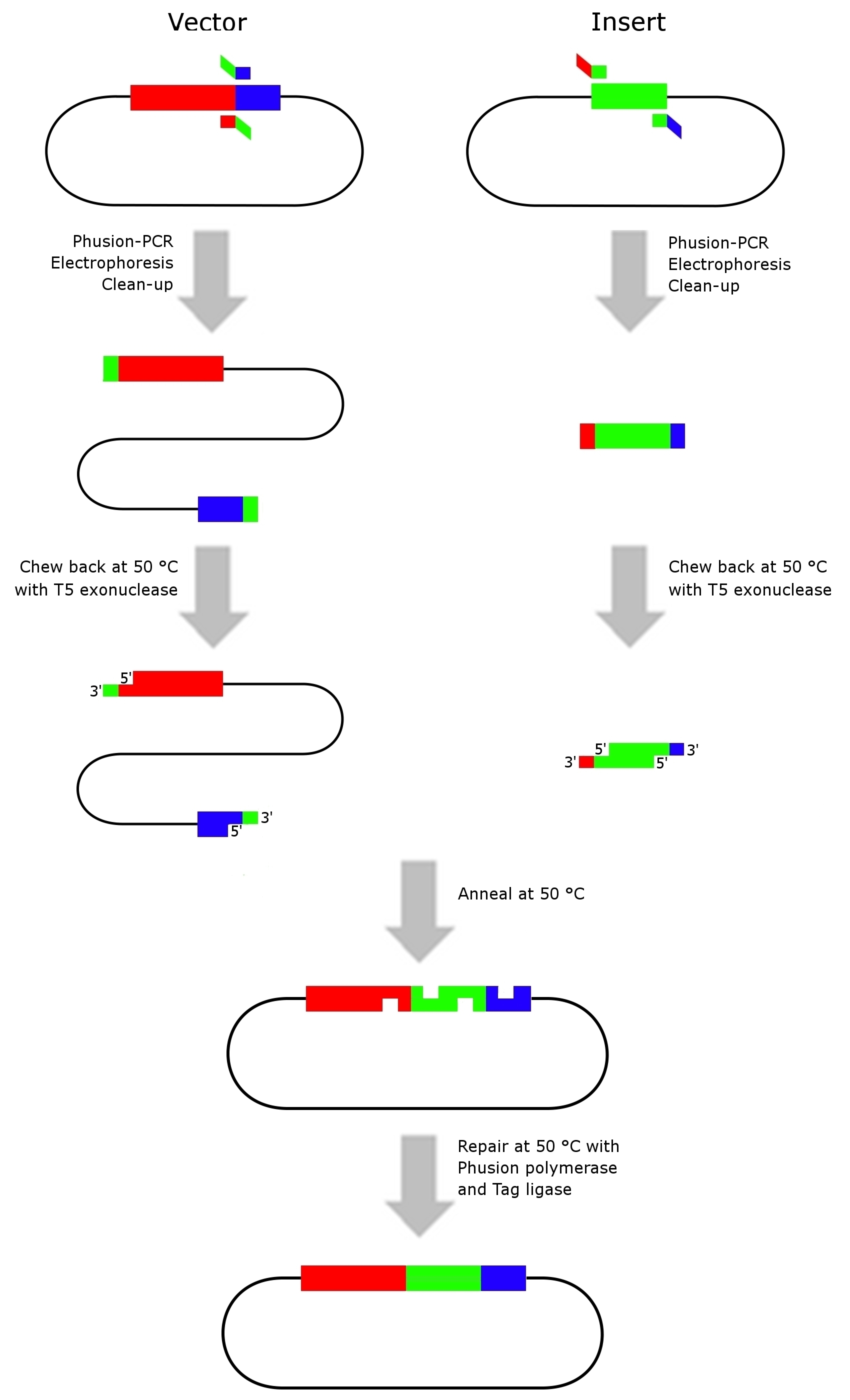
Gibson Assembly
- Modified from [http://www.nature.com/nmeth/journal/v6/n5/full/nmeth.1318.html Gibson et al. (2009)]
- This assembly method is an isothermal, single-reaction method for assembling multiple overlapping DNA molecules. By coordinating the activity of a 5‘ exonuclease, a DNA polymerase and a DNA ligase two adjacent DNA fragments with complementary terminal sequence overlaps can be joined into a covalently sealed molecule, without the use of any restriction endonuclease.
Preparation of DNA molecules for in vitro recombination
- Generate the complementary sequence overlaps by PCR using the Phusion DNA-polymerase. If necessary add 5 M Betain in the reaction mix by reducing the amount of H2O to decrease the number of false PCR products.
- Identify the PCR products of interest by gel electrophoresis with known DNA standards.
- Extract the PCR products from the gel by cutting out the DNA fragments and clean them up by using a commercial PCR clean-up kit.
In vitro recombination
- Assembly mixture
- 320 µL 5x isothermal reaction buffer
- 0.64 µL of 10 U mL-1 T5 exonuclease (for DNA molecules overlapping by greater than 150 bp add 3.2 µL of 10 U ml–1 T5 exonuclease)
- 20 µL of 2 U mL-1 Phusion DNA polymerase
- 160 µL of 40 U mL-1 taq DNA ligase
- add ddH2O water up to a final volume of 1.2 mL
- aliquote 15 µL of the reagent-enzyme mix and store it at –20 ˚C
- Thaw 15 µL assembly mixture aliquot and keep it on ice until use.
- Add 5 µL of the purified DNA molecules in equimolar amounts (between 10 and 100 ng of each DNA fragment).
- Incubate the resulting mixture at 50 ˚C for 15 to 60 min, with 60 min being optimal.
- Transformation (heatshock or Transformation via electroporation) without cleaning up the assembly product.
Restriction analysis
- Digest BioBrick of interest: about 400 ng DNA / 10 µL volume, 1 µL 10x NEBuffer 2.1, 0.5 µL NotI or PstI. Digest for 1 h at 37 °C. NotI is used to determine the length of the BioBrick and the plasmid backbone, PstI is used to determine the length of the BioBrick in the plasmid backbone.
- Gel electrophoresis: add 2 µL loading buffer to every digestion mix, apply about 100 - 200 ng DNA / pocket in gel. Don't forget to apply the uncut BioBrick as well. A good agarose concentration for BioBricks between 0.2 and 3 kb is 1.5 %. The smaller your BioBrick of interest is the higher the agarose concentration should be and vice versa. The gel electrophoresis is made with TAE-buffer. Be sure that you melt your agarose gel in the same buffer you use for the electrophoresis later.
Colony PCR
- Pick one colony with a sterile tip and elute it in 100 µL ddH20 or medium
- Store the colony in 4 °C while colony PCR is running
- One reaction mix contains:
- 10 µL 5x buffer
- 2 µL MgCl2 (25 mM stock)
- 1 µL dNTPs
- 0.5 µL primer mix (prefix/suffix primers or sequencing primers)
- 35.25 µL ddH2O
- 0.25 µL GoTaq polymerase (Promega)
- 1 µL template
- PCR program:
- Start: 3 min, 98 °C
- 30 cycles of:
- 30 s, 98 °C
- 30 s, 55 °C
- 30 s / 1 kb template, 72 °C
- Finish: 5 min, 72 °C
- Gel electrophoresis: check the fragment size
- Plate the correct colony
Analytics
- Protocols for the analytical methods we used are listed in the next chapter.
- DNA and proteins are very small and cannot be seen by the naked eye. To control the success and the results of the upstream and downstream processes, analytical methods are necessary that give reliable results to make DNA or proteins in any way visible for you. The analytical methods we used in our project can be found in this section.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
- This analytical method can be used for separation and identification of proteins according to their electrophoretic mobility. The mobility is a function of length of the molecular weight. Proteins that have identical charge per unit mass due to binding of SDS results in an equal electrophoretic mobility.
Pouring the polyacrylamide gel
- Make a master mix for the stacking and separating gel without adding ammonium persulfate and TEMED.
- Aliquote 6,5 mL for each separating and 2,5 mL for each stacking gel.
- Add ammonium persulfate and TEMED to each separating gel aliquote and pour the solution quickly into the gel casting form. Leave about 2 centimeters below the bottom of the comb for the stacking gel.
- Layer isopropanol on top of the gel.
- Leave the separating gel at room temperature for >60 minutes to polymerize.
- Remove isopropanol and wait until the surface is dry.
- Add ammonium persulfate and TEMED to each stacking gel aliquote and pour the solution quickly into your gel casting form.
- Insert comb without getting bubbles stuck underneath
- Leave the gel at room temperature for >60 minutes to polymerize.
- For storage
- Remove sealing and store the gel wrapped in moistened paper towel at 4°C.
Preparing the sample
- Mix your protein mixture 4:1 with Laemmli-buffer (30 mL protein solution + 10 mL Laemmli-buffer)
- Heat for 5 minutes at 95 °C.
Running the gel
- Remove sealing, put the polymerized gel into gel box and pour SDS running buffer into the negative and positive electrode chamber.
- Remove comp without destroying the gel pocket.
- Pipet the sample into the gel pockets, adjusting the volume according to the amount of protein in your sample. Make sure to include a lane with molecular weight standards (PageRuler Prestained Protein Ladder™ (Fa. Fermentas)) to determinate the molecular weight of the sample.
- Connect the power lead and run the stacking gel with 10 mA until the blue dye front enters the separating gel.
- Raise amperage up to 20 mA for running the separating gel.
- When the distance of the lowest molecular weight standard lane to the gel end is down to 0.5 cm stop the electrophoresis by turning off the power supply.
Polyacrylamide gel staining
Colloidal Coomassie Blue staining
Modified staining protocol from [http://http://newjournal.kcsnet.or.kr/main/j_search/j_abstract_view.htm?code=B021105&qpage=j_search&spage=b_bkcs&dpage=ar Kang et al., 2002].
- agitate the staining solution at 37 °C over night to form the colloids
- After finishing the SDS-PAGE remove gel from gel casting form and transfer it in to a box.
- Add 100 mL of the staining solution to your polyacrylamid gel.
- Incubate the gel in the solution at room temperature until the protein bands got an intensive blue color. Shake the gel continuously during incubation.
- Remove the staining solution
- Wash the gel with dH2O
- Incubate the gel in ddH2O (2-6 h) for bleaching the background. Shake the gel continuously during incubation. If necessary replace the colored water with new one.
MALDI-TOF
- Matrix-assisted Laser Desorption/Ionization – Time of flight (MALDI-TOF) is a procedure to analyze large biomolecules by their mass.
- Before measurement, the analyte is co-crystallized on a metal plate within a solid matrix. 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid) and 2,5-dihydroxybenzoic acid (DHB) are most commonly used.
- Desorption of the analyte is taken by a laser beam, commonly with a wavelength of 337 nm. The matrix absorbs the laser light and the upper layer explosively vaporizes, ejecting both, matrix and analyte molecules. In addition to this vaporization, the laser beam also leads to an ionization of the analytes. The mass analysis is performed by the TOF (time of flight)-analysator. The evaporated ions are accelerated in an electric field. Typical acceleration voltages for this are 10-30 kV. The velocity of an ion depends on its mass and the charge. An ion detector converts the incoming ions into an electrical signal. The advantage of this method is the rapid analysis of a relatively large mass range.
- The MALDI-TOF analysis was performed with the [http://www.bdal.com/products/maldi/ultraflextreme-series/overview.html ultrafleXtreme™] by Bruker Daltonics.
- We used the MALDI-TOF Analysis to characterize the following BioBricks:
- <bbpart>K1172201</bbpart>
- <bbpart>K1172501</bbpart>
- <bbpart>K1172303</bbpart>
Tryptic digest of gel lanes for analysis with MALDI-TOF
- Note:
- Make sure to work under a fume hood.
- Do not work with protective gloves to prevent contamination of your sample with plasticizers.
- Reaction tubes have to be cleaned with 60 % (v/v) CH3CN and 0.1 % (v/v) TFA. Afterwards the solution has to be removed completely followed by evaporation of the tubes under a fume hood. Alternatively microtiter plates from Greiner® (REF 650161) can be used without washing.
- Cut out the protein lanes of a Coomassie-stained SDS-PAGE using a clean scalpel. Gel parts are transferred to the washed reaction tubes/microtiter plate. If necessary cut the parts to smaller slices.
- Gel slices should be washed two times. Therefore add 200 µL 30 % (v/v) acetonitrile in 0,1 M ammonium hydrogen carbonate each time and shake lightly for 10 minutes. Remove supernatant and discard to special waste.
- Dry gel slices at least 30 minutes in a Speedvac.
- Rehydrate gel slices in 15 µL Trypsin-solution followed by short centrifugation.
- Gel slices have to be incubated 30 minutes at room temperature, followed by incubation at 37 °C over night.
- Dry gel slices at least 30 minutes in a Speedvac.
- According to the size of the gel slice, add 5 - 20 µL 50 % (v/v) ACN / 0,1 % (v/v) TFA.
- Samples can be used for MALDI measurement or stored at -20 °C.
- Trypsin-solution: 1 µL Trypsin + 14 µL 10 mM NH4HCO3
- Therefore solubilize lyophilized Trypsin in 200 µL of provided buffer and incubate for 15 minutes at 30 °C for activation. For further use it can be stored at -20 °C.
Preparation and Spotting for analysis of peptides on Bruker AnchorChips
- Spot 0,5 - 1 µL of sample aliquot
- Add 1 µL HCCA matrix solution to the spotted sample aliquots. Pipet up and down approximately five times to obtain a sufficient mixing. Be careful not to contact the AnchorChip.
- Note: Most of the sample solvent needs to be gone in order to achieve a sufficiently low water content. When the matrix solution is added to the previously spotted sample aliquot at a too high water content in the mixture, it will result in undesired crystallization of the matrix outside the anchor spot area.
- Dry the prepared spots at room temperature
- Spot external calibrants on the adjacent calibrant spot positions. Use the calibrant stock solution (Bruker’s “Peptide Calibration Standard II”, Part number #222570), add 125 µL of 0.1% TFA (v/v) in 30% ACN to the vial. Vortex and sonicate the vial.
- Mix the calibrant stock solution in a 1:200 ratio with HCCA matrix and deposit 1 µL of the mixture onto the calibrant spots.
Substrate HPLC
- This method has been used for measurement of the substrate concentration
- Protocol:
- Centrifugation (10 min at 14000 g) of the cell culture
- Mix 900 μL of the supernatant with 2 μL 100 g/L sodium azide solution
- Store the sample at - 20 °C or direct measurement with HPLC
- HPLC parameters:
- Stationary phase: 300 mm x 7.8 mm NUCLEOGEL SUGAR 810H column ([http://www.mn-net.com/HPLCStart/SpecialHPLCphases/Sugars/NUCLEOGELSUGAR810/tabid/10328/language/en-US/Default.aspx Macherey Nagel])
- Column temperature: 60 °C.
- Eluent: 2.5 mM H2SO4-solution
- Flow: 0.8 mL min-1
- Sample volume: 20 μL with a retention time of 25 min
- Detection: RI detector
- Analysis: Software Chromstar 32 version 6.3 ([http://chromstaranalytical.com/index.htm Chromstar Analytical])
NADH-Assay
- This method has been used for measurement of intracellular NADH concentration.
- Protocol:
- Inoculate an overnight culture (30 mL with 1 mL of pre-culture)
- Centrifugate (5 min at 5000 g) of 6-10 mL overnight culture (OD 4-6), adjust volume between different samples for the approximation of the number of cells
- Discard supernatant and wash pellet 3 times with 1 mL of PBS buffer
- Resuspend pellet in 1 mL PBS buffer
- Cell disruption by ribolysation (3 x 30 sec at 6500 rpm)
- Centrifugation for 10 min at maximum speed
- Store supernatant at - 20 ° C or direct measurement with [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]
- [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader] parameters:
- Sample volume = 100 μL clear supernatant
- Excitation = 340 nm
- Emission = 460 nm
- Concentration calculation by NADH calibration curve
Hexadecan Assay
- This assay has been used to measure cell membrane hydrophobicity.
- Protocol
- Inoculate an overnight culture (30 mL with 1 mL of pre-culture)
- Centrifugate (5 min at 4000 g) of 2 mL overnight culture (OD 4-6)
- Discard supernatant and wash pellet 3 times with 1 mL of PBS buffer
- Resuspend pellet in 1 mL 0,9% NaCl and measurement of OD600
- Add x μL of washed cells to 0,9% NaCl for a final volume of 3 mL . Final OD600 should be approximately 0,3 (denoted as A0, calculate exact value)
- Add 3 mL Hexadecan and vortex for 60 sec
- Incubation for 15 min
- Discard the upper organic phase and measure OD600 of the aqueous phase (denoted as A)
- Hydrophobicity can be calculated using the equation: affinity [%] = 100 x [1 – (A/A0)]
Preparing samples for Atomic Force Microscopy (AFM) analysis of the outer membrane of cells
- AFM: During the last years, atomic force microscopy has been used increasingly to investigate microbial surfaces at high resolution. The technique provides three-dimensional images of the surface ultrastructure with molecular resolution, in real time, under physiological conditions, and with minimal sample preparation.
AFM was carried out using the [http://www.bruker.com/de/products/surface-analysis/atomic-force-microscopy/multimode-8/overview.html MultiMode® 8 AFM from Bruker]. The measurements were performed on air with ‘Tapping Mode’ and in water with ‘Peak Force Mode’.
- Cell preparation:
- Inoculating an overnight culture (30 mL with 1 mL of pre-culture) and induction of gene expression if necessary at OD 600 = 1.0.
- Centrifugation (5 min at 5000 g) of 2 mL overnight culture (OD 600 5 ~ 1 ∙ 109 cells/mL).
- Discard supernatant and wash pellet 3 times with 1 mL of ultra pure water.
- Resuspend pellet in 2 mL ultra pure water (2 ∙ 109 cells).
- Preparing of AFM glass slides (0.5 cm2):
- 10 minutes sonication of glass slides in acetone.
- 10 minutes sonication of glass slides in ethanol.
- 10 minutes sonication of glass slides in ultra pure water.
- Dry glass slides under gentle N2 flow.
- Coating of AFM glass slides with prepared cells:
- Drop 5 μL of a 1:4 dilution of prepared cells (1,25 ∙ 106 cells) to the prepared glass slides for a submonolayer coverage.
- Dry glass surface with cells under gentle N2 flow.
- Wash glass surface three times with ultra pure water and dry between under gentle N2 flow.
- The coated glass slides were kept cool until AFM measurement.
NPN membrane permeability assay
- Cell preparation:
- Inoculate an overnight culture (30 mL with 1 mL of pre-culture).
- Centrifugation (8 min at 3000 g) of the whole overnight culture and discard supernatant.
- Resuspend pellet in equal volume of 10 mM PBS buffer.
- Repeat steps 2) and 3) twice.
- Mix 1 mL of washed cells with a 10 mM stock solution of NPN (1 N phenylnaphthylamine) to a final concentration of 10 µM.
- Transfer 200 µL of samples to a flat bottomed black 96 well plate.
- Direct fluorescence measurement with [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader].
- [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader] parameters:
- Sample volume = 200 μL
- Excitation = 355 nm
- Emission Bandwidth = 280 – 550 nm
ONPG membrane permeability assay
- Cell preparation:
- Inoculate an overnight culture (30 mL with 1 mL of pre-culture).
- Centrifugation (8 min at 3000 g) of the whole overnight culture and discard supernatant.
- Resuspend pellet in equal volume of 10 mM PBS buffer.
- Repeat steps 2) and 3) twice.
- Inside a clear, flat bottomed 96 well plate, mix 180 µL ONPG (o-nitrophenyl-b-galactopyranoside) stock solution (3 mM in 10 mM PBS buffer) with 20 µL of washed cells.
- Direct absorbance measurement with [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader].
- [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader] parameters:
- Sample volume = 200 μL
- Absorbance = 405 nm
Cold osmotic shock
- Release of periplasmic protein fraction from E. coli by cold osmotic shock
- Modified protocol from [http://www.jbc.org/content/240/9/3685.full.pdf+html?sid=4a90c176-0ec3-489f-8c82-4734274cebf5 Neu & Heppel, 1965].
- Centrifuge E. coli cell suspension for 5 min at 14,000 g (4 °C) to collect the cells
- Discard the entire supernatant
- Resuspend the cells in ice-cold cell fractionating buffer 1. The resulting volume should be 1/4 of the former suspension volume
- Incubate for 20 min on ice. Invert the suspension at regular intervals to counteract sedimentation
- Centrifuge the cell suspension for 15 min at 14,000 g (4 °C)
- Discard the entire supernatant
- Resuspend the cells in ice-cold cell fractionating buffer 2. The resulting volume should be 1/4 of the former suspension volume
- Incubate for 10 up to 20 min on ice under regular invertion
- Centrifuge the cell suspension for 15 min at 14,000 g (4 °C)
- Save the supernatant, which contains the periplasmatic proteins and membrane proteins when using Cell fractionating buffer 2.2 and 2.3
- If the periplasmatic protein fraction is turbid, re-centrifuge and filter it through a 0.2 µm filter
Riboflavin analysis
HPLC-Method
- Procedure:
- Riboflavin was separated from disturbing substances on a C18 column and detected with a fluorescence detector.
- Elution:
- The sample was injected onto a C18-reversed phase column and eluted with a buffer consisting of 99 % 0.01 M sodium acetate pH 7.4 and 1 % tetrahydrofurane. Total flow is set to 1 ml/min
- Detection:
- Excitation wavelength of 436 nm and emission wavelength of 535 nm
- C18 reverse phase column
- Isocratic method: 99 % 0.01 M sodium acetate pH 7.4 + 1 % tetrahydrofurane
- Flow = 1 mL min-1
- UV-detection at 535 nm
- Internal standard: 5.31 x 10^-5 mg L-1 Riboflavin
- Column:
- Eurospher II 100-5 C18p by Knauer
- Dimensions: 150 x 4.6 mm with precolumn
- Particle size: 5 µm
- Pore size: 100 Å
- Material: silica gel
- Software:
- Clarity (Version 3.0.5.505) by Data Apex
- Autosampler:
- Midas by Spark Holland
- Tray cooling: 10 °C
- Pump:
- L-6200A Intelligent Pump by Hitachi
- UV-Detector:
- Series 1050 by Hewlett Packard
LC-ESI-qTOF-MS(-MS)
HPLC method
- Column: C18 reversed phase (Cogent [http://www.mtc-usa.com/diamond_hydride_specs.asp Diamond Hydride])
- dimension: 150 x 2,1 mm
- Pore size: 100 Å
- Particle size: 4 µm
- Flow: 0.4 mL min-1
- Column temperature: 40 °C
- Injection: 5 µl
- Prerun: 5 min
- Eluent:
- A = 50 % acetonitrile + 50 % H2O + 0,1 % formic acid
- B = 90 % acetonitrile + 10 % H2O + 0,1 % formic acid
- Step gradient:
- 0:00 min: 0 % eluent A
- 8:00 min: 100 % eluent A
- 13:00 min: 100 % eluent A
- 15:30 min: 0 % eluent A
- 18:00 min: 0 % eluent A
- VWR Hitachi LaChrom ULTRA HPLC equipment
- Software: HyStar 3.2, mircrOTOF Control, DataAnalysis 4.0
Ionization method
- Using Bruker Daltonics ESI-QTOF micrOTOFQ
- ESI-QTOF-MS in positive mode
- Mass range: 50 - 1500 m/z
- Source:
- End plate offset: - 500 V, 107 nA
- Capillary: - 3500 V, 4 nA
- Nebulizer: 2 bar
- Dry gas: 8 L min-1
- Dry Temp.: 180 °C
- Transfer:
- Funnel 1 RF: 200 Vpp
- Funnel 2 RF: 300 Vpp
- ISCID Energy: 0 eV
- Heyapole RF: 80 Vpp
- Quadrupole
- Ion energy: 4 eV
- Low mass: 50 m/z
- Collision Cell:
- Collision energy: 8 eV
- Collision RF: 130 Vpp
- Transfer time: 75 µs
- Pre puls storage: 4 µs
MS-MS
- Isolated mass: 243.1 +/- 0.1
- Collision energy: 30 eV
Redox activity of cytochromes via absorption spectroscopy
- Fractioning by cold osmotic shock
- Dilution until measured spectrum is within the visible range
- Exposure to air to facilitate oxidation of the cytochromes
- Measurement of oxidized samples
- Addition of a few grains of sodium dithionite
- Wait for approx. 10min
- Measurement of reduced samples
- Comparison of characteristic peaks
- Peak shifts oxidized to reduced
- Soret band: 410nm → 420nm
- β- and α-bands :broad, undefined peak → 525nm and 552nm
- Measurements were performed with the [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]
Cultivation
- These are the protocols for the cultivations and the downstream processing.
Cultivation Method for UC Davis Collaboration
- Plasmid-DNA of the Anderson promoters J23101, J23108, J23109 and J23111-J23119 was rehydrated in 10 µL ddH2O
- Plasmid-DNA was transformed into competent cells of E.coli-strain DH5α using transformation protocol
- Each part was cultivated in LB-medium containing 20 mg/L chloramphenicol to an OD700 of 0.5 at 37°C with 120 rpm orbital shaking
- Strain control was cultivated under the same condutions without chloramphenicol
- Parts were diluted to a starting OD700 of 0.01 in 200 µL LB and transferred into a clear, flat-bottomed 96-well plate
- Wells were covered by a gas-permeable foil
- The microplate reader(Tecan infinite 200) was ran with the following settings
- Read duration: 18 hours
- Read frequency: Once every ten minutes
- Orbital shaking, amplitude 2.5 mm, frequency 244.5 rpm
- Absorbance measurement at 700 nm
- Fluorescence measurement at 588 nm excitation wavelength and 615 nm emission wavelength
Cultivation Method for Riboflavin Measurements in supernatans
- Most importantly, all cultivations regarding riboflavin production where carried out in M9 medium.
- One ml of a glycerol culture was poured in 30 ml of M9 medium. Cells were grown overnight.
- 100 ml of M9 medium were inoculated with an adjusted amount of overnight culture, so that the starting cell density was identical in different shaking flasks.
- Two samples a one ml were transferred to 1,5 ml microcentrifuge tubes.
- The samples were spun down for 5 min at maximum speed. Afterwards the supernatant was transferred to fresh microcentrifuge tubes.
Materials
- Chemicals, enzymes and kits we used in our lab work.
- Chemical and biological reactions need defined conditions to work as expected. The chemicals, enzymes, kits, buffers and media we used in our project are listed in this section.
Used enzymes
| Enzyme | Producer |
|---|---|
| DpnI | [http://www.fermentas.de/product_info.php?info=p296 Fermentas] |
| EcoRI | NEB |
| GoTaq DNA-polymerase | [http://www.promega.com/products/pcr/routine-pcr/gotaq-pcr-core-systems/ Promega] |
| KOD Hotstart DNA-polymerase | [http://www.merck-chemicals.com/germany/life-science-research/kod-hot-start-dna-polymerase/EMD_BIO-71086/p_iFCb.s1O874AAAEj2Bl9.zLX Novagen] |
| OneTaq DNA-polymerase | [http://www.neb.com/nebecomm/products/productM0480.asp NEB] |
| Pfu DNA-polymerase | [http://www.promega.com/products/pcr/routine-pcr/pfu-dna-polymerase/ Promega] |
| PstI | NEB |
| Phusion HF DNA-polymerase | [http://www.finnzymes.com/pcr/phusion_high_fidelity_dna_polymerase.html Finnzymes] |
| Antarctic phosphatase | NEB |
| SpeI | NEB |
| T4-DNA-Ligase | NEB |
| T5 exonuclease | [http://www.neb.com/nebecomm/products/productM0363.asp NEB] |
| XbaI | NEB |
Used Kits
| Function | Name |
|---|---|
| Plasmid purification | [http://www.fermentas.de/product_info.php?info=p874 Thermo Scientific GeneJET™ Plasmid Miniprep Kit] |
| Plasmid purification | [http://www.promega.de/resources/protocols/technical-bulletins/0/wizard-plus-sv-minipreps-dna-purification-system-protocol Promega Wizard® Plus SV Minipreps DNA Purification System] |
| Plasmid purification | [http://www.analytik-jena.de/de/life-science/produkte/prod/cat/isolierung-von-plasmid-dna/prod/innuprep-plasmid-mini-kit.html Analytik Jena Biometra innuPREP Plasmid Mini Kit] |
| Plasmid purification | [http://www.qiagen.com/products/catalog/sample-technologies/dna-sample-technologies/plasmid-dna/qiaprep-spin-miniprep-kit#orderinginformation Qiagen QIAprep Spin Miniprep Kit] |
| PCR Cleanup | [http://www.mn-net.com/tabid/10745/default.aspx Macherey Nagel NucleoSpin® Extract II] |
| PCR Cleanup | [http://www.promega.com/products/dna-and-rna-purification/dna-fragment-purification/wizard-sv-gel-and-pcr-clean_up-system/ Promega Wizard® SV Gel and PCR Clean-Up] |
| PCR and Gel Cleanup | [http://www.mn-net.com/tabid/1452/default.aspx Macherey Nagel NucleoSpin® Gel and PCR Clean-up] |
| PCR and Gel Cleanup | [http://www.promega.de/products/dna-and-rna-purification/dna-fragment-purification/wizard-sv-gel-and-pcr-clean_up-system/?origUrl=http%3a%2f%2fwww.promega.com%2fproducts%2fdna-and-rna-purification%2fdna-fragment-purification%2fwizard-sv-gel-and-pcr-clean_up-system%2f Promega Wizard® SV Gel and PCR Clean-up System] |
| PCR and Gel Cleanup | [http://www.analytik-jena.de/de/life-science/produkte/prod/cat/cleanup-produkte/prod/innuprep-gel-extraction-kit.html Analytik Jena Biometra innuPREP Gel Extraction Kit] |
| PCR and Gel Cleanup | [http://www.thermoscientificbio.com/nucleic-acid-purification/genejet-gel-extraction-kit Thermo Scientific GeneJET Gel Extraction Kit] |
| PCR core system | [http://www.promega.com/products/pcr/routine-pcr/gotaq-pcr-core-systems/ Promega GoTaq® PCR Core System I] |
Media, buffer, solutions
Chloramphenicol stock solution
- Solubilize 20 mg mL-1 Chloramphenicol in 100 % Ethanol
- Store at -20 °C
Ampicillin stock solution
- Solubilize 100 mg mL-1 Ampicillin
- Store at -20 °C
DNA loading buffer
- 50 % (v/v) glycerol
- 1 mM EDTA
- 0.1 % (w/v) bromphenol blue
- Solve in ddH2O
LB medium
- 10 g Trypton
- 5 g yeast extract
- 10 g NaCl
- 12 g Agar-Agar (for plates)
- Adjust pH to 7.4
SOC medium
- Add the following components for 900 ml of distilled H2O:
- 20 g Trypton
- 5 g Bacto Yeast Extract
- 2 mL of 5 M NaCl
- 2.5 ml of 1 M KCl
- 10 ml of 1 M MgCl2
- 10 ml of 1 M MgSO4
- 20 ml of 1 M glucose
- Adjust to 1L with distilled H2O. Sterilize by autoclaving.
Biotin 500x Stock
- Dissolve 20 mg biotin in 100 mL of 0.05 M NaOH solution and filter sterilize
- Store at 4 °C
- Durable for one year
TAE-Buffer
- 1 L of 50x TAE buffer
- 242.48 g Tris
- 41.02 g sodium acetate
- 18.612 g EDTA
- Adjust pH to 7.8 with acetic acid
- Solve in dH2O
- Dilute 20 mL 50x stock in 1L dH2O for 1x Buffer for PAGE
Cell Fractioning Buffers
CFB 1 (pH 8)
- 0.2 M Tris
- 200 g L-1 sucrose
- 0.1 M EDTA
CFB 2.1 (pH 8)
- 0.01 M Tris
- 0.005 M MgSO4
CFB 2.2 (pH 8)
- 0.01 M Tris
- 0.005 M MgSO4
- 1% Trition
- 2% SDS
CFB 2.3 (pH 8)
- 0.01 M Tris
- 0.005 M MgSO4
- 1% Trition
- 0.2% SDS
CFB 2.4 (pH 8)
- 0.01 M Tris
- 0.005 M MgSO4
- 1% Trition
- 1% SDS
SDS-PAGE gel
The following amounts are for one gel. Stacking gel 5 %:
- 775 μL H2O
- 1.25 mL 0,25 M Tris (pH 6,8)
- 425 μL Bis/Acrylamide (0,8 %, 30 %)
- 50 μL 5 % SDS
- 25 μL 10 % Ammonium persulfate
- 3 μL TEMED
Separating gel 12 %:
- 1.5 mL H2O
- 2.8 mL 1 M Tris (pH 8,8)
- 3.0 mL Bis/ Acrylamide (0,8%, 30%)
- 150 μL 5% SDS
- 37.5 μL 10% Ammonium persulfate
- 5 μL TEMED
SDS running buffer
- 25 mM Tris [pH 8,3]
- 192 mM Glycerol
- 0.1 % SDS
4x Laemmli-buffer
- 250 mM Tris-HCl
- 40 % [v/v] Glycerol
- 20 % [v/v] 2-Mercapthoethanol
- 80 g L-1 SDS
- 0.04 g L-1 BPB
Colloidal Coomassie Brilliant Blue G-250 staining solution
- for 1 L staining solution
- dissolve 50 g L-1 (NH4)2SO4 in ddH2O
- add 10 % (v/v) ethanol
- dissolve 0.2 g L-1 Coomassie Brilliant Blue G-250
- add 2 % (v/v) phosphoric acid
- fill up to 1 L with ddH2O
5x isothermal reaction buffer for Gibson assembly
- storage -20˚C
- 3 mL of 1 M Tris-HCl (pH 7.5)
- 150 µL of 2 M MgCl2,
- 60 µL of 100 mM dGTP
- 60 µL of 100 mM dATP
- 60 µL of 100 mM dTTP,
- 60 µL of 100 mM dCTP
- 300 µL of 1 M DTT
- 1.5 g PEG-8000 and
- 300 µL of 100 mM NAD
Geobacter Medium
- 1.5 g NH4Cl2
- 0.6 g Na2HPO4
- 0.1 g KCl
- 0.82 g Na-acetate
- 10 mL 100 X Trace element solution
- 10 mL 100 X Vitamin solution
- 1 mL 1000 X Selenite-tungstate solution
- 8 g Na-fumarate
- 2.5 g NaHCO3
- 980 mL Distilled water
- 100 X Trace element solution
- 1.5 g Nitrilotriacetic acid
- 3 g MgSO4 x 7 H2O
- 0.5 g MnSO4 x H2O
- 1 g NaCl
- 0.1 g FeSO4 x 7 H2O
- 0.18 g CoSO4 x 7 H2O
- 0.1 g CaCl2 x 2 H2O
- 0.18 g ZnSO4 x 7 H2O
- 0.01 g CuSO4 x 5 H2O
- 0.02 g KAl(SO4)2 x 12 H2O
- 0.01 g H3BO3
- 0.01 g Na2MoO4 x 2 H2O
- 0.03 g NiCl2 x 2 H2O
- 0.3 mg Na2SeO3
- 1000 mL Distilled water
- First dissolve nitrilotriacetic acid and adjust pH to 6.5 with KOH, then add minerals and adjust final pH to 7.0 with KOH.
- 100 X Vitamin Solution
- 2 mg Biotin
- 2 mg Folic acid
- 10 mg Pyridoxine-HCl
- 5 mg Thiamine-HCl x 2 H2O
- 5 mg Riboflavin
- 5 mg Nicotinic acid
- 5 mg D-Ca-pantothenate
- 0,1 mg Vitamin B12
- 5 mg p-Aminobenzoic acid
- 5 mg Lipoic acid12
- 1000 mL Distilled water
- 1000 X Selenite-tungstate solution
- 0,5 g NaOH
- 3 mg Na2SeO3 x 5 H2O
- 4 mg Na2WO4 x H2O
- 1000 mL Distilled water
- Dissolve all ingredients, except fumarate and NaHCO3, bring to a boil, cool down to room temperature while gassing the medium with N2 and add the NaHCO3. Add filter sterilized medium and 0.5 mL/10mL medium from an anoxic filter sterilized 16 % w/v Na-fumarate stock solution to autoclaved cultivation-tubes by injecting it through the silicone stoppers, using a second cannula to prevent overpressure.
M9 Medium
- To prepare 1 liter of M9 minimal medium add the following components to 836.7 mL sterile distilled water. To avoid precipitation, start with CaCl2 and mix the solution thoroughly after addition of a component.
- 100 mL 10 X M9 salt solution
- 50 mL 20 X carbon source
- 10 mL 100 X trace element solution
- 1 mL autoclaved 1 M MgSO4
- 0.3 mL autoclaved 1 M CaCl2
- 1 mL filter sterilized 1 g/L biotin
- 1 mL filter sterilized 1 g/L thiamin
- 10X M9 salt stock solution
- 75.2 g Na2HPO4 x 2H2O
- 30 g KH2PO4
- 5 g NaCl
- 5 g NH4Cl
- Dissolve the salts in 800 mL water and adjust the pH to 7.2 with NaOH. Add water to a final volume of 1 L and autoclave for 15 minutes at 121 °C.
- 20X Carbon source stock solution
| carbon source - | Concentration g/L | Concentration mol/L | Concentration molC/L | Final concentration mmol/L |
|---|---|---|---|---|
| glucose | 200 | 1,11 | 6,66 | 55,55 |
| glycerol | 204,46 | 2,22 | 111 | |
| NA-acetate | 273,15 | 3,33 | 166,5 |
- To produce a 20X stock solution for a single carbon source dissolve the corresponding amount of the carbon source in distilled water and autoclave for 15 minutes at 121 °C.
- 100X trace element solution
- 5 g EDTA
- 0,83 g FeCl3 x 6 H2O
- 84 mg ZnCl2
- 13 mg CuCl2 x 2 H2O
- 10 mg CoCl2 x 6 H2O
- 10 mg H3BO3
- 1,6 mg MnCl2 x 4 H2O
- Dissolve 5g EDTA in 800 ml water and adjust the pH to 7.5 with NaOH. Then add the other components and add distilled water to a final volume of 1 L.
- Sterilize the solution over a 0.22-μm filter
 "
"