Helping Lambert High School
From 2013.igem.org
(Created page with "{{:Team:GeorgiaTech/template}} =Helping Lambert High School= In 2006, the UCSF iGEM team identified the promoter for the hybB subunit of hydrogenase 2 as a cold shock promoter. T...") |
(→The Experiment) |
||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:GeorgiaTech/template}} | {{:Team:GeorgiaTech/template}} | ||
=Helping Lambert High School= | =Helping Lambert High School= | ||
| - | In 2006, the UCSF iGEM team identified the promoter for the hybB subunit of hydrogenase 2 as a cold shock promoter. The team used microarray data as well as their own screening protocol | + | The Lambert High School Team approached us for help with characterizing a previously submitted BioBrick. |
| - | SP: ggaattcgcggccgcttctagagTCTACGTTGCGCTTTTTTATGGCCGG | + | ==Introduction== |
| - | ASP: TTATAGTTATTCCGTTGCGAAGACCTGGC | + | In 2006, the UCSF iGEM team identified the promoter for the hybB subunit of hydrogenase 2 as a cold shock promoter. The team used microarray data as well as their own screening protocol Found here: [https://2006.igem.org/wiki/index.php/UCSF_2005 UCSF 2005] in order to identify cold and heat shock promoters. From our own literature searches, as well as from the results of the characterizations of the hybB promoter by K.U.Leuven and Groningen 2011 iGEM teams, we hypothesize that the hybB promoter was misclassified as cold shock promoter. |
| + | |||
| + | ==Sequence Analysis== | ||
| + | In their conclusions, the Groningen team suggested that the part entered into the registry was too large to be the hybB promoter. We isolated the hybB promoter by first locating the BioBrick part in the BW2952 genome of E. coli using BLAST in the ncbi database. We then isolated the promoter sequence and designed primers to amplify only the promoter (see below lower case indicated BioBrick standard sequence): | ||
| + | SP: ggaattcgcggccgcttctagagTCTACGTTGCGCTTTTTTATGGCCGG | ||
| + | ASP: TTATAGTTATTCCGTTGCGAAGACCTGGC | ||
| + | ==The Experiment== | ||
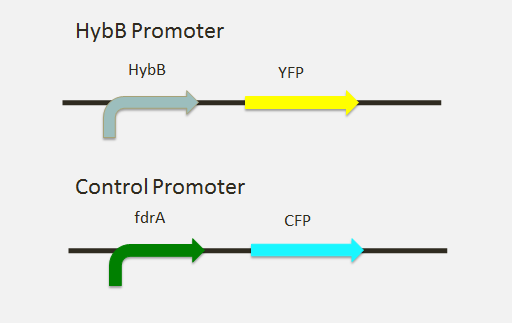
| + | [[File:Lambert_project.PNG|right]] | ||
A fusion with YFP, was created with the promoter using overlap extension PCR as well as Gibson Assembly. We also noticed that many of the previous experiments lacked a control for the thermosensitivity of fluorescent proteins themselves. Therefore we designed a control, using microarray data to identify another promoter that regulated its gene similarly to hybB at room temperature. From this we created a fusion of the fdrA promoter with CFP to be used in characterization as a control. | A fusion with YFP, was created with the promoter using overlap extension PCR as well as Gibson Assembly. We also noticed that many of the previous experiments lacked a control for the thermosensitivity of fluorescent proteins themselves. Therefore we designed a control, using microarray data to identify another promoter that regulated its gene similarly to hybB at room temperature. From this we created a fusion of the fdrA promoter with CFP to be used in characterization as a control. | ||
| - | As a result of time constraints, we were unable to obtain conclusive data using the fluorescent signals. So far we have not been able to find any mention of hybB as a cold shock promoter. | + | As a result of time constraints, we were unable to obtain conclusive data using the fluorescent signals. So far we have not been able to find any mention of hybB as a cold shock promoter. The following paper has some good background on the subject: [http://ac.els-cdn.com/S1369527499800319/1-s2.0-S1369527499800319-main.pdf?_tid=a3428aec-e060-11e2-918b-00000aab0f6b&acdnat=1372471823_d3abbe1cd56f101ccafe19f1acbf71ae Cold-shock response] |
Latest revision as of 03:37, 29 October 2013

Main Page

Contents |
Helping Lambert High School
The Lambert High School Team approached us for help with characterizing a previously submitted BioBrick.
Introduction
In 2006, the UCSF iGEM team identified the promoter for the hybB subunit of hydrogenase 2 as a cold shock promoter. The team used microarray data as well as their own screening protocol Found here: UCSF 2005 in order to identify cold and heat shock promoters. From our own literature searches, as well as from the results of the characterizations of the hybB promoter by K.U.Leuven and Groningen 2011 iGEM teams, we hypothesize that the hybB promoter was misclassified as cold shock promoter.
Sequence Analysis
In their conclusions, the Groningen team suggested that the part entered into the registry was too large to be the hybB promoter. We isolated the hybB promoter by first locating the BioBrick part in the BW2952 genome of E. coli using BLAST in the ncbi database. We then isolated the promoter sequence and designed primers to amplify only the promoter (see below lower case indicated BioBrick standard sequence):
SP: ggaattcgcggccgcttctagagTCTACGTTGCGCTTTTTTATGGCCGG ASP: TTATAGTTATTCCGTTGCGAAGACCTGGC
The Experiment
A fusion with YFP, was created with the promoter using overlap extension PCR as well as Gibson Assembly. We also noticed that many of the previous experiments lacked a control for the thermosensitivity of fluorescent proteins themselves. Therefore we designed a control, using microarray data to identify another promoter that regulated its gene similarly to hybB at room temperature. From this we created a fusion of the fdrA promoter with CFP to be used in characterization as a control. As a result of time constraints, we were unable to obtain conclusive data using the fluorescent signals. So far we have not been able to find any mention of hybB as a cold shock promoter. The following paper has some good background on the subject: [http://ac.els-cdn.com/S1369527499800319/1-s2.0-S1369527499800319-main.pdf?_tid=a3428aec-e060-11e2-918b-00000aab0f6b&acdnat=1372471823_d3abbe1cd56f101ccafe19f1acbf71ae Cold-shock response]
 "
"