Team:UGent/Project
From 2013.igem.org
| Line 15: | Line 15: | ||
<html> | <html> | ||
<body> | <body> | ||
| + | <br> | ||
<p>Therefore a new method was developed for the overexpression of a gene of interest in the bacterial chromosome: Chemically Inducible Chromosomal evolution (CIChE). In this technique the chromosome is evolved to contain a higher number of gene copies by adding a chemical inducer. The original model for CIChE, however, results in bacterial strains containing a large number of antibiotic resistance genes.</p> | <p>Therefore a new method was developed for the overexpression of a gene of interest in the bacterial chromosome: Chemically Inducible Chromosomal evolution (CIChE). In this technique the chromosome is evolved to contain a higher number of gene copies by adding a chemical inducer. The original model for CIChE, however, results in bacterial strains containing a large number of antibiotic resistance genes.</p> | ||
| - | {{:Team:UGent/Templates/ToggleBoxStart}} Read more about CIChE{{:Team:UGent/Templates/ToggleBoxStart1}}{{:Team:UGent/Templates/ToggleBoxStart2}}In 2009, Tyo et al. developed a technique for the stable, high copy expression of a gene of interest in E. coli without the use of high copy number plasmids, thus avoiding their previously stated negative characteristics. They called this plasmid-free, high gene copy expression system ‘chemically inducible chromosomal evolution’. In this method, the gene of interest is integrated in the microbial genome and then amplified to achieve multiple copies and reach the desired expression level. Genomic integration guarantees ordered inheritance, resolving the problem of allele segregation. | + | </body> |
| + | </html> | ||
| + | {{:Team:UGent/Templates/ToggleBoxStart}} Read more about CIChE{{:Team:UGent/Templates/ToggleBoxStart1}}{{:Team:UGent/Templates/ToggleBoxStart2}} | ||
| + | <html> | ||
| + | <body> | ||
| + | <p>In 2009, Tyo et al. developed a technique for the stable, high copy expression of a gene of interest in E. coli without the use of high copy number plasmids, thus avoiding their previously stated negative characteristics. They called this plasmid-free, high gene copy expression system ‘chemically inducible chromosomal evolution’. In this method, the gene of interest is integrated in the microbial genome and then amplified to achieve multiple copies and reach the desired expression level. Genomic integration guarantees ordered inheritance, resolving the problem of allele segregation. | ||
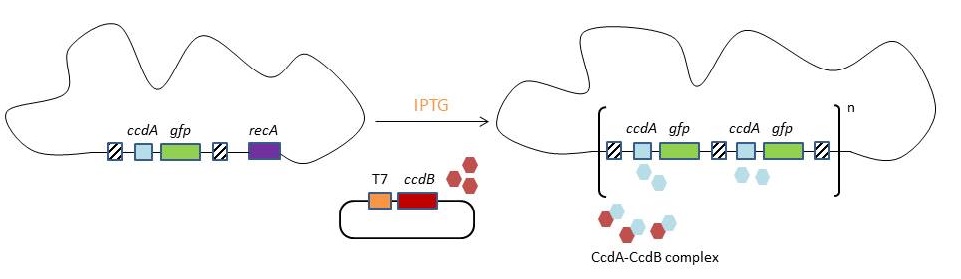
CIChE works as follows: First a construct, containing the gene(s) of interest and the antibiotic marker chloramphenicol acetyl transferase (''cat'') flanked by homologous regions, is delivered to and subsequently integrated into the ''E. coli'' genome. The construct can be amplified in the genome through tandem gene duplication by recA homologous recombination. Then the strain is cultured in increasing concentrations of chloramphenicol, providing a growth advantage for cells with increased repeats of the construct and thereby selecting for bacteria with a higher gene copy number ('''Figure'''). | CIChE works as follows: First a construct, containing the gene(s) of interest and the antibiotic marker chloramphenicol acetyl transferase (''cat'') flanked by homologous regions, is delivered to and subsequently integrated into the ''E. coli'' genome. The construct can be amplified in the genome through tandem gene duplication by recA homologous recombination. Then the strain is cultured in increasing concentrations of chloramphenicol, providing a growth advantage for cells with increased repeats of the construct and thereby selecting for bacteria with a higher gene copy number ('''Figure'''). | ||
| - | [[File:UGent_2013_CIChE.jpg|thumb|400px|center|Tyo et al., 2009]]This process is called chromosomal evolution. When the desired gene copy number is reached, ''recA'' is deleted, thereby fixing the copy number.{{:Team:UGent/Templates/ToggleBoxEnd}}To make this valuable technique more widely applicable in the industry, we developed a model for chromosomal evolution based on a toxin-antitoxin system instead of antibiotic resistance. | + | [[File:UGent_2013_CIChE.jpg|thumb|400px|center|Tyo et al., 2009]]This process is called chromosomal evolution. When the desired gene copy number is reached, ''recA'' is deleted, thereby fixing the copy number.</p> |
| - | + | </body> | |
| - | {{:Team:UGent/Templates/ToggleBoxStart}} Read more about antibiotic resistance{{:Team:UGent/Templates/ToggleBoxStart1}}{{:Team:UGent/Templates/ToggleBoxStart2}}Antibiotic resistance is the resistance of a bacterium to an antibacterial medicine a.k.a. antibiotic to which it was originally sensitive. When a pathogenic bacterium has acquired resistance to a certain antibiotic, infections of this bacterium can no longer be treated with the antibiotic in question since the bacterium has become insensitive to the antibiotic. | + | </html> |
| + | {{:Team:UGent/Templates/ToggleBoxEnd}} | ||
| + | <html> | ||
| + | <body> | ||
| + | <p>To make this valuable technique more widely applicable in the industry, we developed a model for chromosomal evolution based on a toxin-antitoxin system instead of antibiotic resistance.</p> | ||
| + | </body> | ||
| + | </html> | ||
| + | {{:Team:UGent/Templates/ToggleBoxStart}} Read more about antibiotic resistance{{:Team:UGent/Templates/ToggleBoxStart1}}{{:Team:UGent/Templates/ToggleBoxStart2}} | ||
| + | <html> | ||
| + | <body> | ||
| + | <p>Antibiotic resistance is the resistance of a bacterium to an antibacterial medicine a.k.a. antibiotic to which it was originally sensitive. When a pathogenic bacterium has acquired resistance to a certain antibiotic, infections of this bacterium can no longer be treated with the antibiotic in question since the bacterium has become insensitive to the antibiotic. | ||
Overuse and misuse of antibiotics has accelerated the emergence and spread of resistant bacteria. Today antibiotic resistance has become a fairly well-known term as a result of the recurring appearance of multi-drug resistant bacteria such as MRSA and VRE in media, awareness campaigns etc. These bacteria have become resistant to the commonly used antibiotics. As a result infections are very difficult to treat. The emergence of these hard-to-kill pathogenic bacteria poses a serious risk to public health. | Overuse and misuse of antibiotics has accelerated the emergence and spread of resistant bacteria. Today antibiotic resistance has become a fairly well-known term as a result of the recurring appearance of multi-drug resistant bacteria such as MRSA and VRE in media, awareness campaigns etc. These bacteria have become resistant to the commonly used antibiotics. As a result infections are very difficult to treat. The emergence of these hard-to-kill pathogenic bacteria poses a serious risk to public health. | ||
| - | In biotechnology antibiotic resistance genes (the genes that make a bacterium resistant to a certain antibiotic) are often used as selectable markers. The best known application is probably the selection of plasmids. In the original CIChE technique a chloramphenicol resistance gene is used as selectable marker. During chromosomal evolution the antibiotic resistance gene is duplicated since it is the driving pressure for the tandem duplication of the CIChE-construct. As a result the final bacterial strain carries multiple antibiotic resistance genes. The creation of such a strain raises several safety concerns as bacteria can pass resistance genes on to other, possibly pathogenic, bacteria in the environment through a process called horizontal gene transfer. Although this is very unlikely to occur the possibility of horizontal gene transfer cannot be entirely ruled out. We will try to replace the use of antibiotic resistance genes with a toxin-antitoxin system. By eliminating the need for antibiotic resistance genes, CIChE could be applied in the industry without having to worry about the possibility of horizontal gene transfer and the spread of antibiotic resistance. {{:Team:UGent/Templates/ToggleBoxEnd}} | + | In biotechnology antibiotic resistance genes (the genes that make a bacterium resistant to a certain antibiotic) are often used as selectable markers. The best known application is probably the selection of plasmids. In the original CIChE technique a chloramphenicol resistance gene is used as selectable marker. During chromosomal evolution the antibiotic resistance gene is duplicated since it is the driving pressure for the tandem duplication of the CIChE-construct. As a result the final bacterial strain carries multiple antibiotic resistance genes. The creation of such a strain raises several safety concerns as bacteria can pass resistance genes on to other, possibly pathogenic, bacteria in the environment through a process called horizontal gene transfer. Although this is very unlikely to occur the possibility of horizontal gene transfer cannot be entirely ruled out. We will try to replace the use of antibiotic resistance genes with a toxin-antitoxin system. By eliminating the need for antibiotic resistance genes, CIChE could be applied in the industry without having to worry about the possibility of horizontal gene transfer and the spread of antibiotic resistance.</p> |
| + | </body> | ||
| + | </html> | ||
| + | {{:Team:UGent/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | <html> | ||
| + | <body> | ||
| - | + | <h1>Our model</h1> | |
| - | + | <h1>Toxin-Antitoxin systems</h1> | |
| - | In our opinion it is possible to eliminate the need for antibiotics by using a toxin-antitoxin system. These systems are widely distributed among bacteria and archaea. The toxins produced by these systems can slow down or stop cell growth by interfering with certain molecules that are essential in cellular processes like DNA replication, cell wall synthesis, ATP synthesis etc.. Under normal conditions, the toxin is inhibited by the antitoxin, which is encoded in the same operon as the toxin. Research revealed different types of TA systems. | + | <p>In our opinion it is possible to eliminate the need for antibiotics by using a toxin-antitoxin system. These systems are widely distributed among bacteria and archaea. The toxins produced by these systems can slow down or stop cell growth by interfering with certain molecules that are essential in cellular processes like DNA replication, cell wall synthesis, ATP synthesis etc.. Under normal conditions, the toxin is inhibited by the antitoxin, which is encoded in the same operon as the toxin. Research revealed different types of TA systems.</p> |
| + | </body> | ||
| + | </html> | ||
{{:Team:UGent/Templates/ToggleBoxStart}} Read more about the different types of TA systems {{:Team:UGent/Templates/ToggleBoxStart1}}{{:Team:UGent/Templates/ToggleBoxStart2}} | {{:Team:UGent/Templates/ToggleBoxStart}} Read more about the different types of TA systems {{:Team:UGent/Templates/ToggleBoxStart1}}{{:Team:UGent/Templates/ToggleBoxStart2}} | ||
''' Type I''' | ''' Type I''' | ||
Revision as of 19:44, 10 August 2013
|

Tweets van @iGEM_UGent |
|
|
 "
"