Team:DTU-Denmark/Notebook/24 July 2013
From 2013.igem.org
| Line 46: | Line 46: | ||
*'Nir' or 'pZ', tells which product is being made | *'Nir' or 'pZ', tells which product is being made | ||
*'1' - part 1 of Nir OR '2' - part 2 of Nir OR 'w' - whole Nir (on the pZ tubes 1 and 2 are just duplicates) | *'1' - part 1 of Nir OR '2' - part 2 of Nir OR 'w' - whole Nir (on the pZ tubes 1 and 2 are just duplicates) | ||
| - | *'5' - used 5uL of N.europeae culture as template OR '10' - used 10uL of N.europeae culture as template | + | *'5' - used 5uL of ''N. europeae'' culture as template OR '10' - used 10uL of N.europeae culture as template |
* 'ex' - extraction PCR, using the new non-USER primers that Jakob made OR 'U' - using the new USER primers that Jakob made | * 'ex' - extraction PCR, using the new non-USER primers that Jakob made OR 'U' - using the new USER primers that Jakob made | ||
Revision as of 10:31, 22 August 2013
24 July 2013
Contents |
208
Main purpose
- Make USER-reaction to get pZA21 with arabinose inducible promoter.
- Make PCR to get more HAO and AMO with USER-primers.
- Finish the restriction analysis.
Who was in the lab
Henrike, Julia, Gosia, Kristian
Procedure
USER reaction and transformation
We perform USER reaction, samples are as follows:
- plasmid pZA21 and AMO
- plasmid pZA21 and HAO
- negative control with water instead of insert
Reaction is performed in the same way as on 18-07-2013 with increased amount of insert (up to 14 uL due to low DNA concentration).
With the same procedure we also preformed a USER reaction with pZA21::RFP PCR-amplified with no promoter and araBAD from BBa_K808000.
Restriction analysis
From last week transformants with AMO and HAO in pZA21 we performed plasmid isolation.
We perform restriction analysis with EcoRI. Expected fragments are as follows:
- For pZA21 with AMO:
3309 pz, 2283 pz
- For pZA21 with HAO:
2233 pz, 2124 pz, 930 pz
Primer diluting
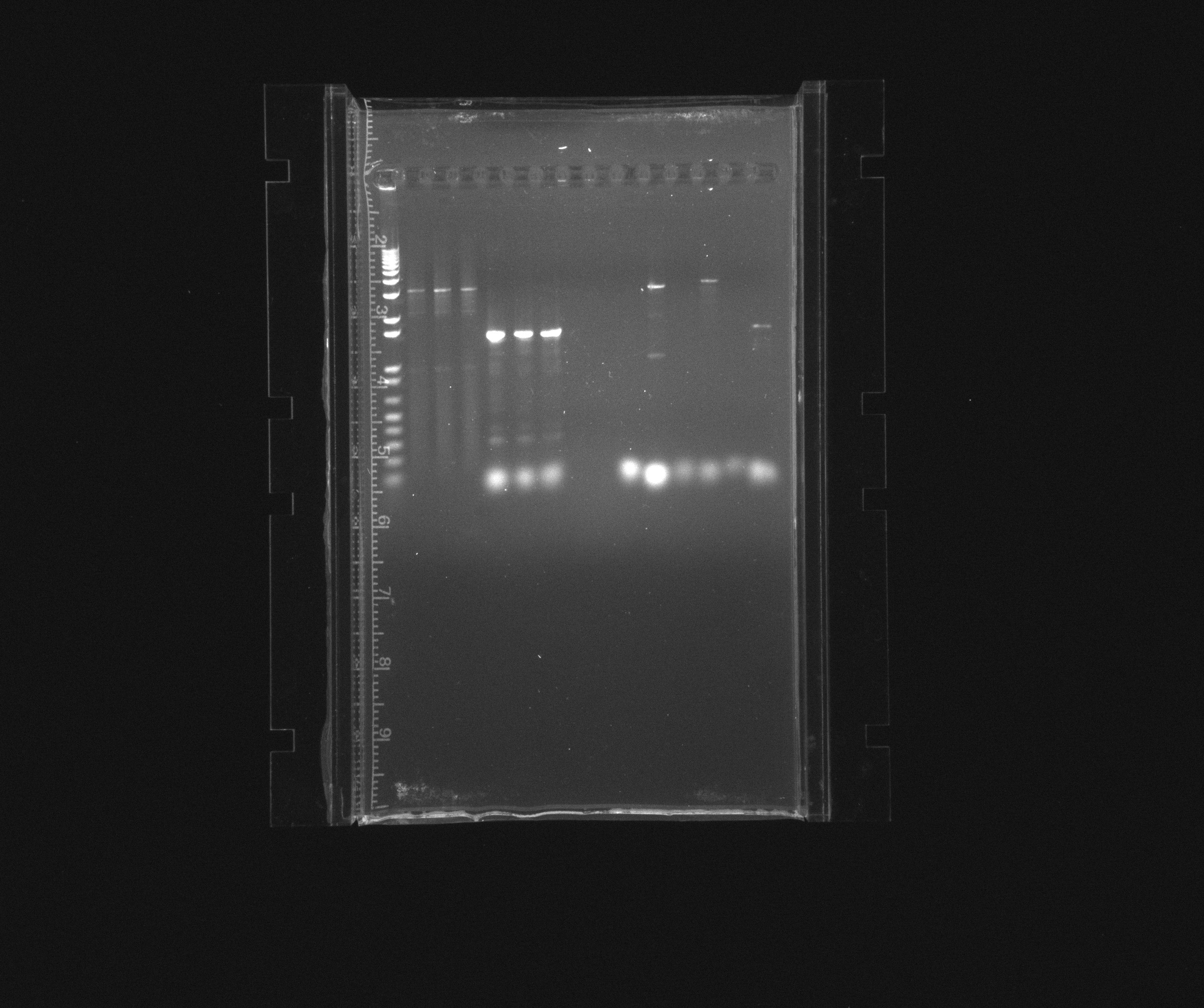
PCR to extract Nir from Pseudosomonas with new USER primers
Tubes have 4 labels (pZA21 only has 3), in vertical order:
- 'Nir' or 'pZ', tells which product is being made
- '1' - part 1 of Nir OR '2' - part 2 of Nir OR 'w' - whole Nir (on the pZ tubes 1 and 2 are just duplicates)
- '5' - used 5uL of N. europeae culture as template OR '10' - used 10uL of N.europeae culture as template
- 'ex' - extraction PCR, using the new non-USER primers that Jakob made OR 'U' - using the new USER primers that Jakob made
two programs:
all extraction PCRs and part 2 of Nir with USER primers - 54C, 5:00
part 1 of Nir with USER primers and pZA21 for ligation with Nir - 50C, 5:00
PCR with USER-primers on HAO and AMO
Standard USER-PCR was made with the HAO and AMO templates purified earlier today. The reactions where done in duplicates and with 2 different concentrations:
- HAO 5 is with 5uL template
- HAO 10 is with 10uL template
- AMO 5 is with 5uL template
- AMO 10 is with 10uL template
All tubes where run with 52C annealing temperature and 3 min extension time.
Results
first gel
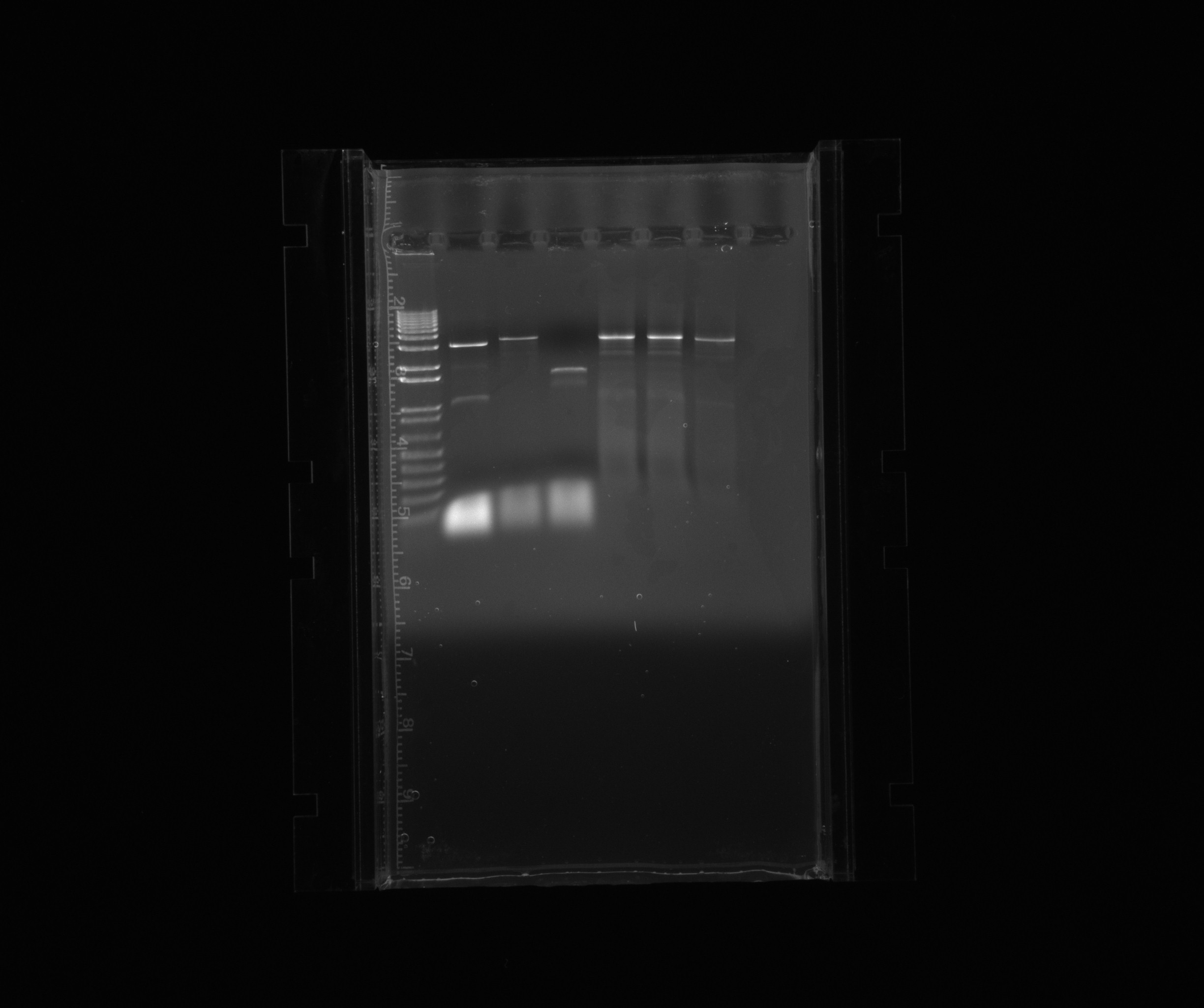
decided to purify RFP in pZA21 without promoter as well as HAO, AMO and cycAX
purification gel
loaded the complete PCR reaction (~45 uL), cut out the bands of our products and used QIAgen gel extraction kit
- 1: 1 kb ladder
- 2: HAO
- 3: AMO
- 4: cycAX
- 5: RPF in pZA21 without promoter
- 6: RPF in pZA21 without promoter
- 7: RPF in pZA21 without promoter
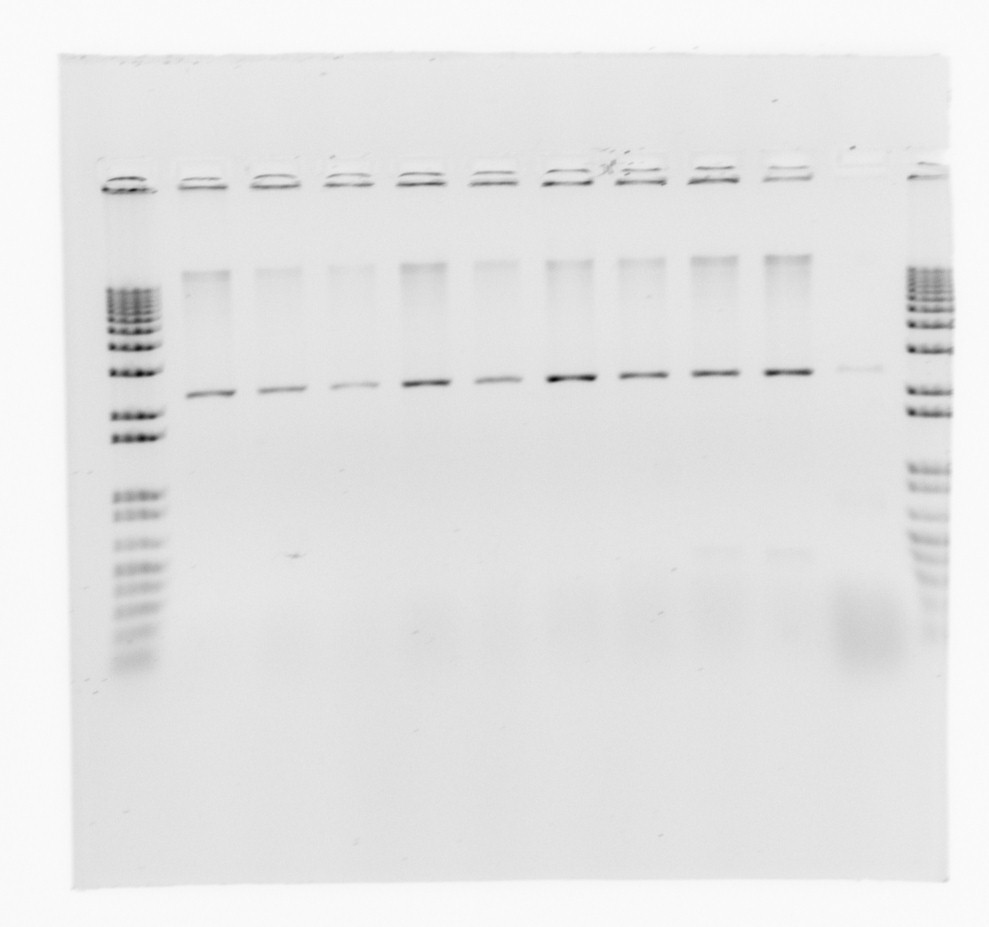
gel for restriction analysis
Conclusion: We only get one band for each plasmid, so the inserts are not present.
Conclusion
No of the constructs, Nir, HAO, AMO was shown to have an insert by the restriction analysis. There was only a faint band for the cycAX construct but this have already been verified by colony PCR yesterday and by the orange color of the colonies. We will make a new restriction analysis tomorrow to see if we can locate the insert. Also we will chech transformant from USER-cloning and possibly make another USER-cloning with the new arabinose inducible vector system.
Navigate to the Previous or the Next Entry
 "
"