Team:uOttawa/Project
From 2013.igem.org
| (6 intermediate revisions not shown) | |||
| Line 61: | Line 61: | ||
</div> | </div> | ||
| - | <div class="row accent"> | + | <div class="row borderTop accent"> |
<div class="container"> | <div class="container"> | ||
<h2>References</h2> | <h2>References</h2> | ||
| - | < | + | <ul class="reference"> |
| - | + | <li>Ellis, Tom, Xiao Wang, and James J Collins. "Diversity-based, model-guided construction of synthetic gene networks with predicted functions". <i>Nature Biotechnology</i> 27.5 (2009): 465-471. Web. 30 May 2013.</li> | |
| - | < | + | <li>Goentoro, Lea, Oren Shoval, Marc W. Kirschner, and Uri Alon. "The Incoherent Feedforward Loop Can Provide Fold-Change Detection in Gene Regulation". <i>Molecular Cell</i> 36 (2009): 894-899. Print.</li> |
| - | + | <li>Mads Kærn and Ron Weiss. "Synthetic gene regulatory systems". In: <i>Systems Modeling in Cellular Biology.</i> Z. Sallasi <i>et al.</i> (Eds.). MIT Press (2006).</li> | |
| - | < | + | <li>Mangan, Shmoolik and Uri Alon. "Structure and function of the feed-forward loop network motif". <i>Proceedings of the National Academy of Sciences</i> 100.21 (2003): 11980–11985. Web. 9 May 2013.</li> |
| - | + | <li>McIsaaca, R. Scott, Sanford J. Silvermana, Megan N. McCleana, Patrick A. Gibneya, Joanna Macinskasa, Mark J. Hickmana, Allegra A. Pettia, and David Botsteina. "Fast-acting and nearly gratuitous induction of gene expression and protein depletion in <i>Saccharomyces cerevisiae</i>". <i>Molecular Biology of the Cell</i> 22 (2011): 4447-4459. Web. 7 May 2013.</li> | |
| - | < | + | </ul> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 16:13, 30 May 2013
Project Description
Background

Figure 1. The type-I incoherent feedforward loop.
The Type-I Incoherent Feedforward Loop and Fold-Change Detection

Figure 1. The type-I incoherent feedforward loop.
The type-I incoherent feedforward loop (I1-FFL) is a gene network in which protein X activates a gene Z while simultaneously activating the production of a repressor of gene Z, labeled Y.
For this year’s project, we are aiming to construct an I1-FFL that can detect fold-changes in the levels of protein X. In other words, the expression of Z would be reliant on the relative change in X as opposed to absolute values of X. For example, if X changes from an arbitrary concentration of 1 to an arbitrary concentration of 5, the level of expression of gene Z would be exactly the same as that resulting from a change from a concentration of 5 to a concentration of 25 of X (a fold-change by a factor of 5 in both cases).
How Fold-Change Detection is Achieved
This fold-change detection is enabled through the repressor Y. Take the case when activator X, repressor Y, and protein Z each have an arbitrary concentration of 1 in the cell (1:1:1 ratio). If the concentration of X is doubled, the ratio between X and Y now becomes 2:1. X immediately activates Z, but since the repressor Y takes time to fold, it lags behind, and the concentration of Z spikes to 2 due to the activation. When Y folds into its active form and carries out its repressive activity, the concentration of Z returns to its initial concentration of 1. This is because at this point, X and Y return to a 1:1 ratio in the cell. In order for the concentration of Z to reach 2 once again, the X:Y ratio must first return to 2:1 - thus, X has to reach a concentration of 4 before the same output of Z is produced. In this way, fold-change detection is achieved.

Figure 2. The fold-change detection schematic using an incoherent feedforward loop. Concentrations of X, Y, and Z are shown in arbitrary units.
Engineering our System
The Design of our Gene Network and the Interactions Between Components
In our proposed design, rtTA will act as our activator (X), Lacl as our repressor (Y), and GFP as our reporter protein (Z). Each part is detectable via a different fluorescent protein. A visual representation of our pathway is shown in Figure 3.
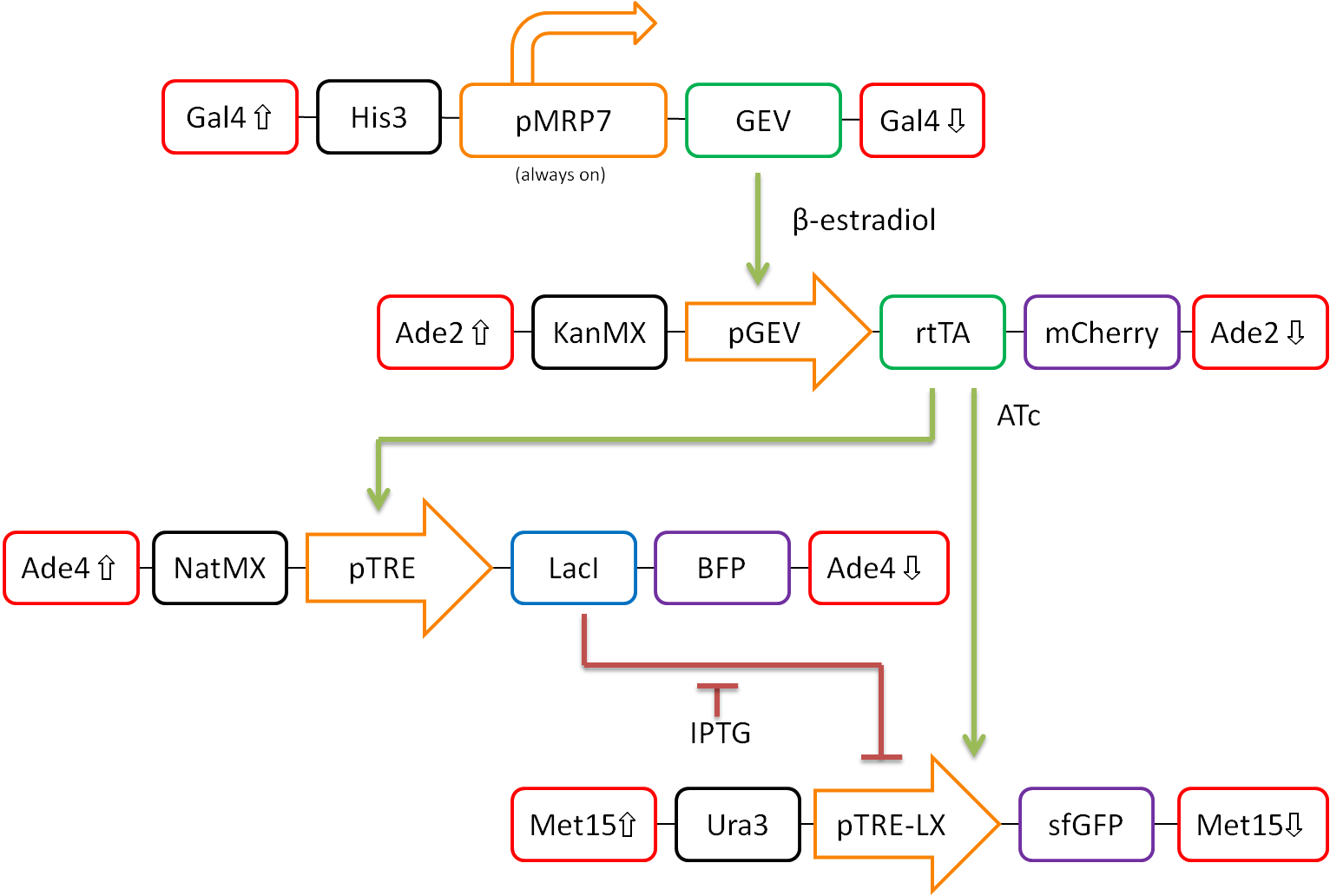
Figure 3. Network design for fold-change detection of varying levels of rtTA using a type-I incoherent feedforward loop. Red boxes indicate regions of the S.cerevisiae genome to be targeted, while black boxes signify selection markers. Orange arrows represent promoters. Green and blue boxes represent activators and repressors respectively, and purple boxes signify varying fluorescent proteins.
This network design allows for calibration of the network via tunable levels of β-estradiol, allowing for accurate control of the amount of rtTA(X) in the cell. This will allow us to ensure that fold-change detection works as theorized in our network.
Furthermore, activation by rtTA is mediated by varying levels of anhydrotetracycline (ATc), and LacI repression is mediated by varying levels of Isopropyl β-D-1-thiogalactopyranoside (IPTG). These methods of mediation allow for high tunability of the system, and we should be able to find a concentration range of ATc and IPTG at which the system will work as desired.
The system will then be characterized extensively. The fluorescent markers tagged to each component of the network will allow for simple temporal analysis using flow cytometry. This will allow for a high-throughput analysis of the activity of our network.
The Potential Application
A Fold-Change Detection System for Toxic Molecules
Once our system is tuned and is working, it can be modified to act as a fold-change detector for toxic molecules. By replacing the pGEV promoter in front of the rtTA gene with a promoter that is inducible by a toxic molecule, the amount of rtTA produced will be directly correlated to the concentration of the toxic molecule in the cellular environment.

Figure 4. Modification of the network illustrated in Figure 3 to enable toxic molecule detection. The pGEV promoter will be exchanged for a promoter responsive to a toxic molecule.
This detector system is advantageous in that it detects fold-changes rather than changes in absolute values, which gives meaning to a signal in reference to the background signal level. This allows for the network to respond only to a signal that rises significantly above the background noise.
References
- Ellis, Tom, Xiao Wang, and James J Collins. "Diversity-based, model-guided construction of synthetic gene networks with predicted functions". Nature Biotechnology 27.5 (2009): 465-471. Web. 30 May 2013.
- Goentoro, Lea, Oren Shoval, Marc W. Kirschner, and Uri Alon. "The Incoherent Feedforward Loop Can Provide Fold-Change Detection in Gene Regulation". Molecular Cell 36 (2009): 894-899. Print.
- Mads Kærn and Ron Weiss. "Synthetic gene regulatory systems". In: Systems Modeling in Cellular Biology. Z. Sallasi et al. (Eds.). MIT Press (2006).
- Mangan, Shmoolik and Uri Alon. "Structure and function of the feed-forward loop network motif". Proceedings of the National Academy of Sciences 100.21 (2003): 11980–11985. Web. 9 May 2013.
- McIsaaca, R. Scott, Sanford J. Silvermana, Megan N. McCleana, Patrick A. Gibneya, Joanna Macinskasa, Mark J. Hickmana, Allegra A. Pettia, and David Botsteina. "Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae". Molecular Biology of the Cell 22 (2011): 4447-4459. Web. 7 May 2013.
 "
"

