Team:DTU-Denmark/Notebook/6 September 2013
From 2013.igem.org
(→Procedure) |
|||
| (11 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:DTU-Denmark/Templates/StartPage|6 September 2013}} | {{:Team:DTU-Denmark/Templates/StartPage|6 September 2013}} | ||
| - | |||
Navigate to the [[Team:DTU-Denmark/Notebook/5_September_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/7_September_2013|Next]] Entry | Navigate to the [[Team:DTU-Denmark/Notebook/5_September_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/7_September_2013|Next]] Entry | ||
=Lab 208= | =Lab 208= | ||
| Line 6: | Line 5: | ||
==Main purpose== | ==Main purpose== | ||
<hr/> | <hr/> | ||
| - | *Add USER endings to HAO gene extracted from ''Nitrosomonas | + | *Add USER endings to HAO gene extracted from ''Nitrosomonas europaea'' |
==Who was in the lab== | ==Who was in the lab== | ||
<hr/> | <hr/> | ||
| - | + | Henrike | |
==Procedure== | ==Procedure== | ||
<hr/> | <hr/> | ||
| - | ===Repeat PCR from yesterday with same conditions and programs=== | + | ===Repeat HAO USER PCR from yesterday with same conditions and programs=== |
Template: HAO extraction fragment -isolated from ''Nitrosomonas europea''- (purified from PCR run on 04.09) | Template: HAO extraction fragment -isolated from ''Nitrosomonas europea''- (purified from PCR run on 04.09) | ||
| Line 48: | Line 47: | ||
*Add 49 ul of the master mix, and | *Add 49 ul of the master mix, and | ||
*Add 1 ul DMSO (for the 5%) or 1 ul of MQ (for the 3%) | *Add 1 ul DMSO (for the 5%) or 1 ul of MQ (for the 3%) | ||
| + | |||
| + | ===User reaction=== | ||
| + | |||
| + | was performed with HAO User fragment that was purified yesterday (but had low concentration) and pZA21::ara (promoter from the Biobrick K808000) and pZA21 with a tight arabinose promoter (from our SPL). Also Nir 1 User and Nir 2 User were ligated into the pZA21 ara tight for Nir. Negative controls were made for all three backbones. | ||
==Results== | ==Results== | ||
<hr/> | <hr/> | ||
| - | == | + | |
| - | + | ===Gel 1=== | |
| + | Gel of yesterday's PCR was empty (we lost the picture though...) | ||
| + | |||
| + | PCR was redone. | ||
| + | |||
| + | ===Gel 2=== | ||
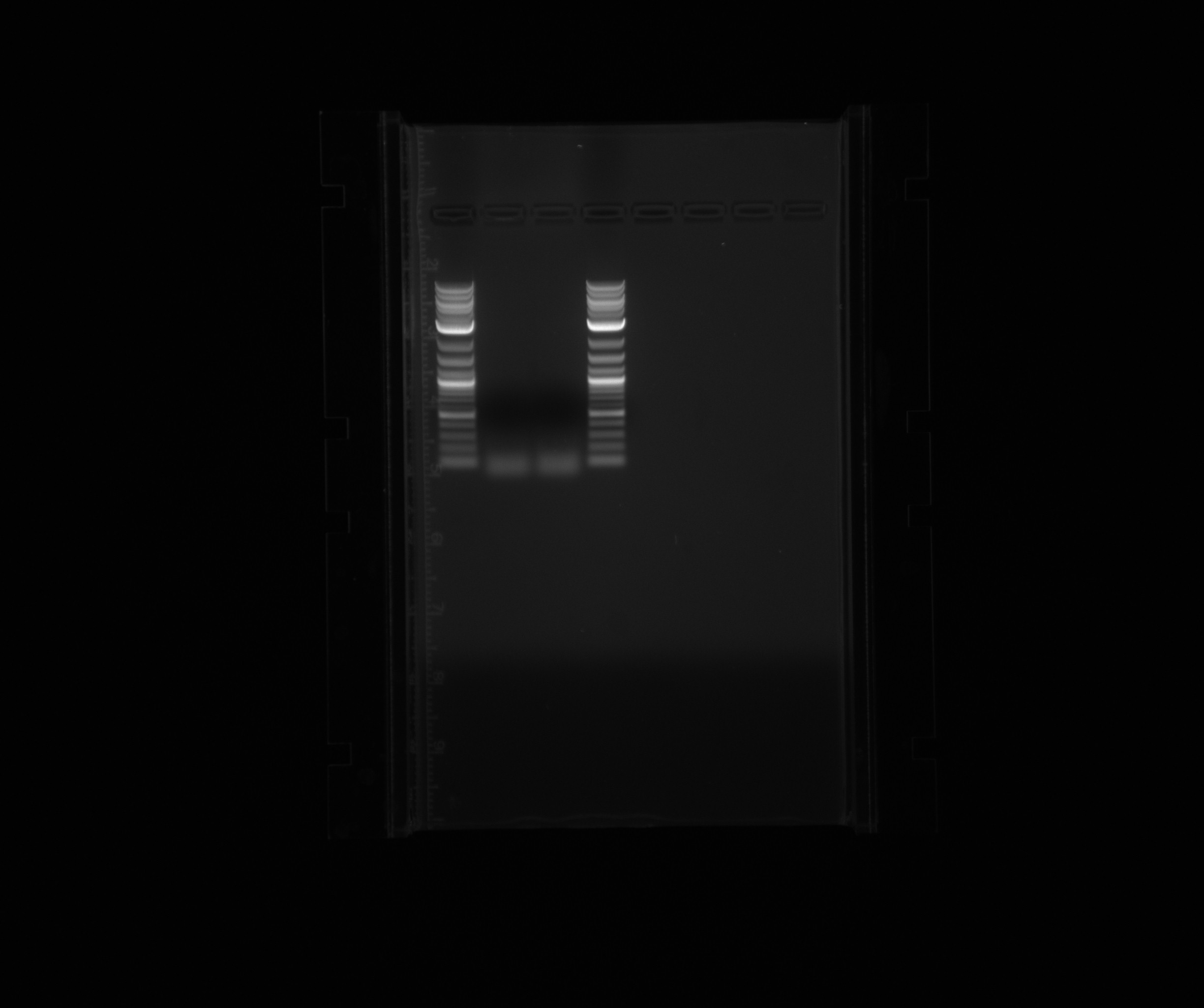
| + | |||
| + | For the PCR results of today. | ||
| + | |||
| + | * 2-log ladder | ||
| + | * HAO User on 59C, 3% DMSO | ||
| + | * HAO User on 59C, 5% DMSO | ||
| + | * 2-log ladder | ||
| + | |||
| + | [[File:2013-09-06 HAO USER 59.jpg|600px]] | ||
| + | |||
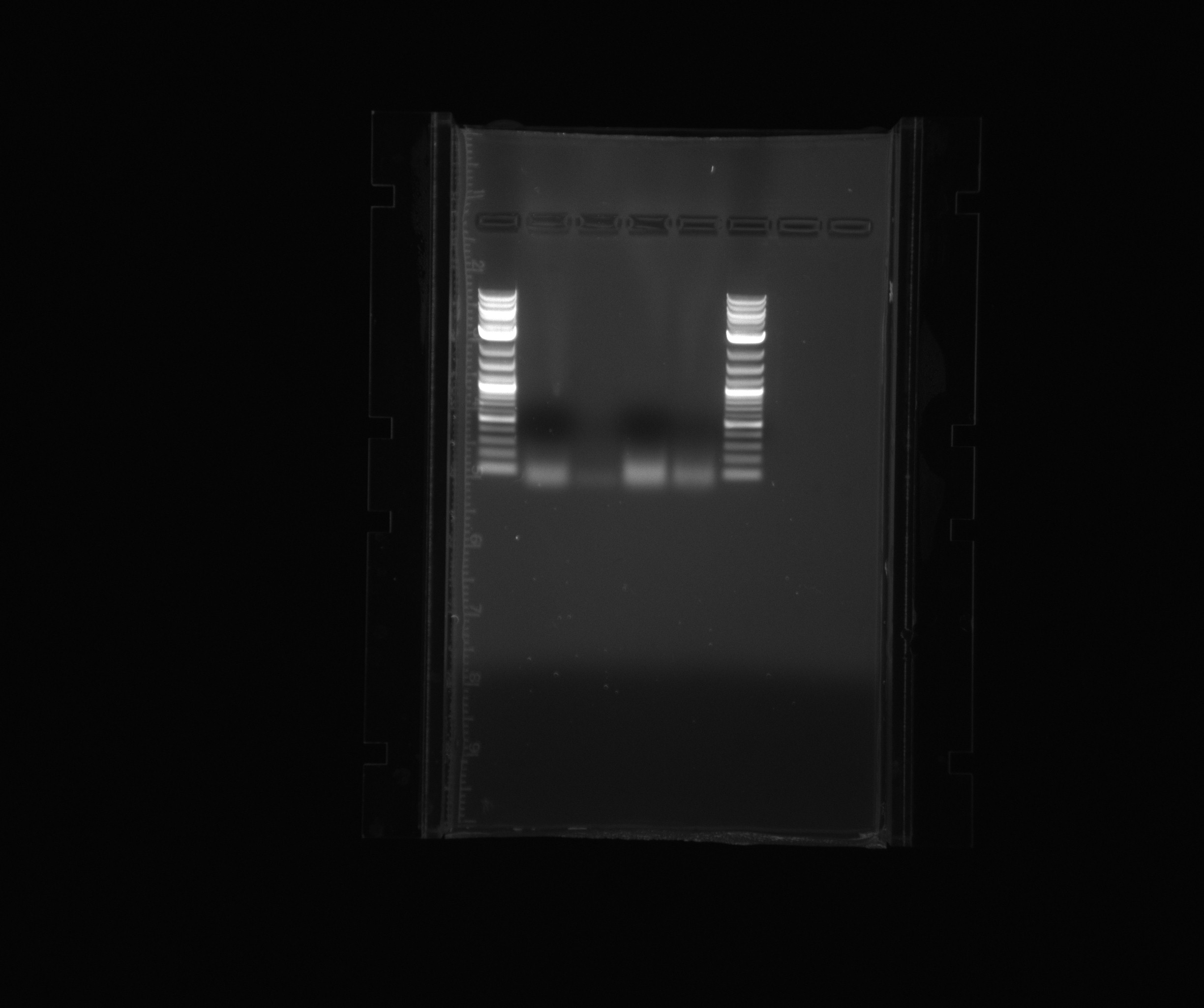
| + | ===Gel 3=== | ||
| + | |||
| + | * 2-log ladder | ||
| + | * HAO User on touchdown, 3% DMSO | ||
| + | * HAO User on touchdown, 5% DMSO | ||
| + | * 2-log ladder | ||
| + | |||
| + | [[File:2013-09-06 HAO USER td.jpg|600px]] | ||
| + | |||
Latest revision as of 10:55, 4 October 2013
6 September 2013
Contents |
Lab 208
Main purpose
- Add USER endings to HAO gene extracted from Nitrosomonas europaea
Who was in the lab
Henrike
Procedure
Repeat HAO USER PCR from yesterday with same conditions and programs
Template: HAO extraction fragment -isolated from Nitrosomonas europea- (purified from PCR run on 04.09)
Primers: 18a & 18b
Two versions, one with 5% DMSO and one with 3% DMSO and two programs: Touchdown (62C -> 55) and a single annealing temperature of 59C.
| compound | volume (uL) |
|---|---|
| dNTPs | 1 |
| HF | 10 |
| polymerase x7 | 0.5 |
| FW | 3 |
| RV | 3 |
| template | 1 |
| MQ | 29 |
| DMSO | 1.5 |
- Add 49 ul of the master mix, and
- Add 1 ul DMSO (for the 5%) or 1 ul of MQ (for the 3%)
User reaction
was performed with HAO User fragment that was purified yesterday (but had low concentration) and pZA21::ara (promoter from the Biobrick K808000) and pZA21 with a tight arabinose promoter (from our SPL). Also Nir 1 User and Nir 2 User were ligated into the pZA21 ara tight for Nir. Negative controls were made for all three backbones.
Results
Gel 1
Gel of yesterday's PCR was empty (we lost the picture though...)
PCR was redone.
Gel 2
For the PCR results of today.
- 2-log ladder
- HAO User on 59C, 3% DMSO
- HAO User on 59C, 5% DMSO
- 2-log ladder
Gel 3
- 2-log ladder
- HAO User on touchdown, 3% DMSO
- HAO User on touchdown, 5% DMSO
- 2-log ladder
Navigate to the Previous or the Next Entry
 "
"