Template:Kyoto/Notebook/Sep 12
From 2013.igem.org
(Difference between revisions)
(→Colony PCR) |
(→Colony PCR) |
||
| (28 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
!DNA||concentration[µg/mL]||260/280||260/230 | !DNA||concentration[µg/mL]||260/280||260/230 | ||
|- | |- | ||
| - | |Pcon- | + | |Pcon-pT181attenuator-DT||293.6 ||1.88 ||1.84 |
|- | |- | ||
|Pcon-apt12-1R-DT||303.8 ||1.75 ||1.64 | |Pcon-apt12-1R-DT||303.8 ||1.75 ||1.64 | ||
|- | |- | ||
| - | | | + | |Fusion1attenuator(pSB1c3)||172.0 ||1.96 ||2.31 |
|- | |- | ||
|RBS-lisis1-DT||235.1||1.59||1.60 | |RBS-lisis1-DT||235.1||1.59||1.60 | ||
| Line 33: | Line 33: | ||
|- | |- | ||
|} | |} | ||
| - | [[File: | + | [[File:igku_9121.png]]<br> |
| + | |||
===Liquid Culture=== | ===Liquid Culture=== | ||
<div class="experiment"> | <div class="experiment"> | ||
| Line 42: | Line 43: | ||
|9/10 Pcon-apt12-1R-DT1||Plusgrow medium(+Amp) | |9/10 Pcon-apt12-1R-DT1||Plusgrow medium(+Amp) | ||
|- | |- | ||
| - | |9/10 DT-Pcon- | + | |9/10 DT-Pcon-pT181attenuator1||Plusgrow medium(+CP) |
|- | |- | ||
| - | |9/10 Fusion1- | + | |9/10 Fusion1-attenuator(pSB1c3)||Plusgrow medium(+CP) |
|} | |} | ||
</div> | </div> | ||
| Line 52: | Line 53: | ||
<span class="author">Tatsui</span></div> | <span class="author">Tatsui</span></div> | ||
| - | |||
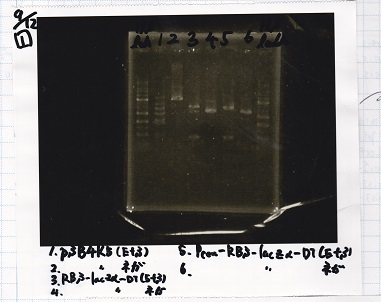
{| class="wikitable" | {| class="wikitable" | ||
!Lane||Sample | !Lane||Sample | ||
| Line 64: | Line 64: | ||
|4||RBS-lacz-DT(EcoRI&SpeI) | |4||RBS-lacz-DT(EcoRI&SpeI) | ||
|- | |- | ||
| - | |5|| | + | |5|| EcoRI&SpeI NC( EcoRI&SpeI) |
|- | |- | ||
|6||Pcon-RBS-lacz-DT( EcoRI&SpeI) | |6||Pcon-RBS-lacz-DT( EcoRI&SpeI) | ||
| Line 74: | Line 74: | ||
|} | |} | ||
| - | [[File: | + | [[File:igku_9122.png]] |
</div> | </div> | ||
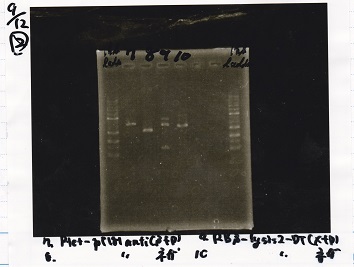
| Line 84: | Line 84: | ||
|1||1kb ladder | |1||1kb ladder | ||
|- | |- | ||
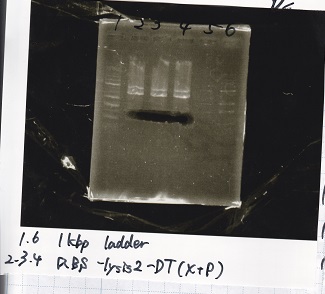
| - | |2||Ptet- | + | |2||Ptet-pT181antisense(SpeI&PstI) |
|- | |- | ||
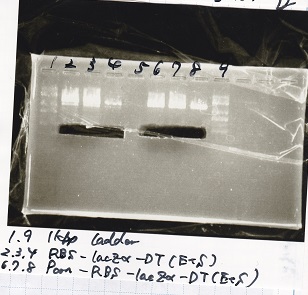
| - | |3|| Ptet- | + | |3|| Ptet-pT181antisense NC(SpeI&PstI) |
|- | |- | ||
|4||RBS-lysis2-DT(XbaI&PstI) | |4||RBS-lysis2-DT(XbaI&PstI) | ||
|- | |- | ||
| - | |5|| | + | |5|| EcoRI&SpeI NC( EcoRI&SpeI) |
|- | |- | ||
|6||-- | |6||-- | ||
| Line 100: | Line 100: | ||
|} | |} | ||
| - | [[File: | + | [[File:igku_9123.png]] |
===Gel Extraction=== | ===Gel Extraction=== | ||
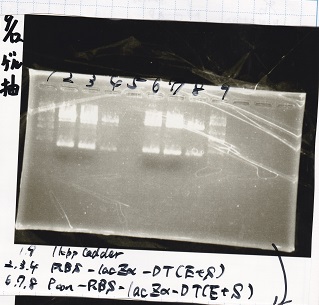
| Line 127: | Line 127: | ||
|} | |} | ||
| - | [[File: | + | [[File:igku_9129.png]]<br> |
| - | [[File: | + | [[File:igku_91210.png]]<br> |
{| class="wikitable" | {| class="wikitable" | ||
!Name||concentration[µg/mL]||260/280||260/230 | !Name||concentration[µg/mL]||260/280||260/230 | ||
| Line 159: | Line 159: | ||
|} | |} | ||
| - | [[File: | + | [[File:igku_9127.png]]<br> |
| - | [[File: | + | [[File:igku_9128.png]]<br> |
{| class="wikitable" | {| class="wikitable" | ||
!Name||concentration[µg/mL]||260/280||260/230 | !Name||concentration[µg/mL]||260/280||260/230 | ||
| Line 184: | Line 184: | ||
|3||Pbad/araC-RBS-RFP(PCR product) | |3||Pbad/araC-RBS-RFP(PCR product) | ||
|} | |} | ||
| - | + | [[File:igku_9124.png]] | |
===Colony PCR=== | ===Colony PCR=== | ||
| Line 248: | Line 248: | ||
===Colony PCR=== | ===Colony PCR=== | ||
<div class="experiment"> | <div class="experiment"> | ||
| - | <span class="author">Nakamoto | + | <span class="author">Nakamoto and Tatsui</span> |
{| class="wikitable" | {| class="wikitable" | ||
!Sample||base pair | !Sample||base pair | ||
|- | |- | ||
| - | |9/11 Pcon- | + | |9/11 Pcon-pT181attenuator-RBS-lacZα-DT||965 |
|- | |- | ||
|9/11 Plux-PBS-lysis1-DT 1||613 | |9/11 Plux-PBS-lysis1-DT 1||613 | ||
| Line 277: | Line 277: | ||
===Transformation=== | ===Transformation=== | ||
<div class="experiment"> | <div class="experiment"> | ||
| - | <span class="author"> | + | <span class="author">Tatsui</span> |
{| class="wikitable" | {| class="wikitable" | ||
!Name||Sample||Competent Cells||Plate | !Name||Sample||Competent Cells||Plate | ||
| Line 310: | Line 310: | ||
|} | |} | ||
| - | [[File: | + | [[File:igku_91211.png]] |
</div> | </div> | ||
| Line 334: | Line 334: | ||
|8|| 1kb ladder | |8|| 1kb ladder | ||
|} | |} | ||
| + | [[File:igku_91212.png]]<br> | ||
===Electrophoresis=== | ===Electrophoresis=== | ||
| Line 368: | Line 369: | ||
|} | |} | ||
</div> | </div> | ||
| + | [[File:igku_91213.png]] | ||
===Liquid Culture=== | ===Liquid Culture=== | ||
| Line 410: | Line 412: | ||
|5min||30s||30s||36s||30cycles | |5min||30s||30s||36s||30cycles | ||
|} | |} | ||
| - | + | ||
<br> | <br> | ||
| Line 419: | Line 421: | ||
!Lane||Sample||Enzyme | !Lane||Sample||Enzyme | ||
|- | |- | ||
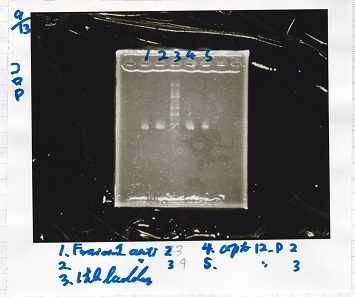
| - | |1||Fusion1 | + | |1||Fusion1 antisense 2||-- |
|- | |- | ||
| - | |2|| Fusion1 | + | |2||Fusion1 antisense 3||-- |
|- | |- | ||
|3||1kb ladder ||-- | |3||1kb ladder ||-- | ||
| Line 430: | Line 432: | ||
|} | |} | ||
</div> | </div> | ||
| + | [[File:igku_91214.png]] | ||
===Restriction Enzyme Digestion=== | ===Restriction Enzyme Digestion=== | ||
<div class="experiment"> | <div class="experiment"> | ||
| - | <span class="author"> | + | <span class="author">Tatsui </span> |
{| class="wikitable" | {| class="wikitable" | ||
| - | ! ||9/12 RBS-lysis2-DT|| | + | ! ||9/12 RBS-lysis2-DT||XbaI||PstI||Buffer||BSA||MilliQ||total |
|- | |- | ||
|2 cuts||5.2 ||1.0µL||1.0µL||3.0µL||3.0µL||16.8µL ||30µL | |2 cuts||5.2 ||1.0µL||1.0µL||3.0µL||3.0µL||16.8µL ||30µL | ||
| Line 443: | Line 446: | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | ! ||9/12 RBS-lysis3-DT|| | + | ! ||9/12 RBS-lysis3-DT||XbaI ||PstI||Buffer||BSA||MilliQ||total |
|- | |- | ||
|2 cuts||5.2 ||1.0µL||1.0µL||3.0µL||3.0µL||16.8µL ||30µL | |2 cuts||5.2 ||1.0µL||1.0µL||3.0µL||3.0µL||16.8µL ||30µL | ||
| Line 451: | Line 454: | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | ! ||9/11 RBS-lysis1-DT|| | + | ! ||9/11 RBS-lysis1-DT||XbaI||PstI||Buffer||BSA||MilliQ||total |
|- | |- | ||
|2 cuts||8.5||1.0µL||1.0µL||3.0µL||3.0µL||13.5µL ||30µL | |2 cuts||8.5||1.0µL||1.0µL||3.0µL||3.0µL||13.5µL ||30µL | ||
| Line 459: | Line 462: | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | ! ||8/20 J23100-RBS-luxR -DT|| | + | ! ||8/20 J23100-RBS-luxR -DT||EcoRI ||Buffer||BSA||MilliQ||total |
|- | |- | ||
|1cut||5.8||1.0µL||3.0µL||3.0µL||17.2µL ||30µL | |1cut||5.8||1.0µL||3.0µL||3.0µL||17.2µL ||30µL | ||
| Line 466: | Line 469: | ||
|} | |} | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | ! ||9/10 pSB1C3|| | + | ! ||9/10 pSB1C3||EcoRI ||SpeI||Buffer||BSA||MilliQ||total |
|- | |- | ||
|2cuts||8.1||1.0µL||1.0µL||3.0µL||3.0µL||13.9µL ||30µL | |2cuts||8.1||1.0µL||1.0µL||3.0µL||3.0µL||13.9µL ||30µL | ||
| Line 474: | Line 477: | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | ! ||9/12 Plux || | + | ! ||9/12 Plux ||PstI ||Buffer||BSA||MilliQ||total |
|- | |- | ||
|1cut||12.0||1.0µL||3.0µL||3.0µL||11.0micro;L ||30µL | |1cut||12.0||1.0µL||3.0µL||3.0µL||11.0micro;L ||30µL | ||
| Line 482: | Line 485: | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | ! ||9/10 pSB1C3|| | + | ! ||9/10 pSB1C3||XbaI ||PstI||Buffer||BSA||MilliQ||total |
|- | |- | ||
|2cuts||8.1||1.0µL||1.0µL||3.0µL||3.0µL||13.9µL ||30µL | |2cuts||8.1||1.0µL||1.0µL||3.0µL||3.0µL||13.9µL ||30µL | ||
| Line 496: | Line 499: | ||
!Lane||Sample||Enzyme1||Enzymel2 | !Lane||Sample||Enzyme1||Enzymel2 | ||
|- | |- | ||
| - | |1||9/12 RBS-lysis2-DT || | + | |1||9/12 RBS-lysis2-DT ||XbaI||PstI |
|- | |- | ||
|2||9/12 RBS-lysis2-DT NC ||--||-- | |2||9/12 RBS-lysis2-DT NC ||--||-- | ||
|- | |- | ||
| - | |3||9/11 RBS-lysis1-DT || | + | |3||9/11 RBS-lysis1-DT ||XbaI||PstI |
|- | |- | ||
|4||9/11 RBS-lysis1-DT NC ||--||-- | |4||9/11 RBS-lysis1-DT NC ||--||-- | ||
|- | |- | ||
| - | |5|| 9/11 RBS-lysis3-DT || | + | |5|| 9/11 RBS-lysis3-DT ||XbaI||PstI |
|- | |- | ||
|6|| 9/11 RBS-lysis3-DT NC1||--||-- | |6|| 9/11 RBS-lysis3-DT NC1||--||-- | ||
| Line 512: | Line 515: | ||
|8|| 100bp ladder||--||-- | |8|| 100bp ladder||--||-- | ||
|- | |- | ||
| - | |9||J23100-RBS-luxR-DT || | + | |9||J23100-RBS-luxR-DT ||EcoRI||-- |
|- | |- | ||
|10|| J23100-RBS-luxR-DT ||--||-- | |10|| J23100-RBS-luxR-DT ||--||-- | ||
|- | |- | ||
| - | |11||9/10 pSB1C3 || | + | |11||9/10 pSB1C3 ||EcoRI||SpeI |
|- | |- | ||
|12|| 9/10 pSB1C3 NC||--||--- | |12|| 9/10 pSB1C3 NC||--||--- | ||
|- | |- | ||
| - | |13||9/10 pSB1C3 || | + | |13||9/10 pSB1C3 ||XbaI||PstI |
|- | |- | ||
| - | |14||9/12 Plux|| | + | |14||9/12 Plux||PstI||-- |
|- | |- | ||
|15||9/12 Plux NC||--||-- | |15||9/12 Plux NC||--||-- | ||
| + | |} | ||
| + | [[File:igku_91215.png]]<br> | ||
| + | </div> | ||
| + | |||
| + | ====Observation through a confocal microscope==== | ||
| + | We used the liquid culture media with 200μL 9/10Pcon-pT181antisense-spinach-DT& 200μL 9/10 Pcon-spinach-DT& 200μL 9/10 JM109(overnight culture). | ||
| + | <br> | ||
| + | We translated them into each 1.5mL tube. <br> | ||
| + | |||
| + | We elminated each supernatant using a 5000xg centrifuge for 1minute,. <br> | ||
| + | Then,we resuspended with 100µL M9(distilled water) and eliminated each supernatant using a 5000xg centrifuge for 1 minute.x2<br> | ||
| + | |||
| + | Adding 100µL M9(in 20µM DFHBI), we resuspended the pret. <br> | ||
| + | |||
| + | 15min after incubating in shield light, we eliminated each supernatant using a 5000xg centrifuge for 1 minute. <br> | ||
| + | |||
| + | We looked at the fluorescence of the pret. | ||
| + | <br> | ||
| + | We failed to observe it. | ||
| + | |||
| + | ===Liquid Culture=== | ||
| + | <div class="experiment"> | ||
| + | <span class="author">Hirano</span> | ||
| + | {| class="wikitable" | ||
| + | !Sample||medium | ||
| + | |- | ||
| + | |9/10 Pcon-pT181antisense-spinach-DT||Plusgrow medium | ||
| + | |- | ||
| + | |9/10 Pcon-spinach-DT||Plusgrow medium | ||
| + | |- | ||
| + | |9/1 spinach-DT(RNA master plate)|| Plusgrow medium | ||
|} | |} | ||
</div> | </div> | ||
Latest revision as of 20:49, 27 September 2013
Sep 12
Miniprep
| DNA | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Pcon-pT181attenuator-DT | 293.6 | 1.88 | 1.84 |
| Pcon-apt12-1R-DT | 303.8 | 1.75 | 1.64 |
| Fusion1attenuator(pSB1c3) | 172.0 | 1.96 | 2.31 |
| RBS-lisis1-DT | 235.1 | 1.59 | 1.60 |
| RBS-lisis2-DT | 387.7 | 1.91 | 2.01 |
| RBS-lisis3-DT | 387.9 | 1.91 | 2.09 |
| Plux | 166.7 | 1.98 | 1.63 |
Electrophoresis
| Lane | Sample |
|---|---|
| 1 | 100bp ladder |
| 2 | 9/11 P-RBS-lisis1-DT(Colony PCR prodution) |
Liquid Culture
| Sample | medium |
|---|---|
| 9/10 Pcon-apt12-1R-DT1 | Plusgrow medium(+Amp) |
| 9/10 DT-Pcon-pT181attenuator1 | Plusgrow medium(+CP) |
| 9/10 Fusion1-attenuator(pSB1c3) | Plusgrow medium(+CP) |
Electrophoresis
| Lane | Sample |
|---|---|
| 1 | 1kb ladder |
| 2 | pSB4K5(EcoRI&SpeI) |
| 3 | pSB4K5 NC(EcoRI&SpeI) |
| 4 | RBS-lacz-DT(EcoRI&SpeI) |
| 5 | EcoRI&SpeI NC( EcoRI&SpeI) |
| 6 | Pcon-RBS-lacz-DT( EcoRI&SpeI) |
| 7 | Pcon-RBS-lacz-DT NC( EcoRI&SpeI) |
| 8 | 1kb ladder |
2
| Lane | Sample |
|---|---|
| 1 | 1kb ladder |
| 2 | Ptet-pT181antisense(SpeI&PstI) |
| 3 | Ptet-pT181antisense NC(SpeI&PstI) |
| 4 | RBS-lysis2-DT(XbaI&PstI) |
| 5 | EcoRI&SpeI NC( EcoRI&SpeI) |
| 6 | -- |
| 7 | -- |
| 8 | 1kb ladder |
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kb ladder | -- |
| 2 | RBS-lacZα-DT | EcoRI&SpeI |
| 3 | RBS-lacZα-DT | EcoRI&SpeI |
| 4 | RBS-lacZα-DT | EcoRI&SpeI |
| 5 | -- | -- |
| 6 | Pcon-RBS-lacZα-DT | EcoRI&SpeI |
| 7 | Pcon-RBS-lacZα-DT | EcoRI&SpeI |
| 8 | Pcon-RBS-lacZα-DT | EcoRI&SpeI |
| 9 | 1kb ladder | -- |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| RBS-lacZα-DT(EcoRI&SpeI) | 9.8 | 1.50 | 0.39 |
| Pcon-RBS-lacZα-DT(EcoRI&SpeI) | 11.2 | 1.47 | 0.08 |
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kb ladder | -- |
| 2 | RBS-lysis2-DT | XbaI&PstI |
| 3 | RBS-lysis2-DT | XbaI&PstI |
| 4 | RBS-lysis2-DT | XbaI&PstI |
| 5 | -- | -- |
| 6 | 1kb ladder | -- |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| RBS-lysis2-DT(XbaI&PstI) | 5.4 | 1.43 | 0.77 |
Electrophoresis
| Lane | Sample |
|---|---|
| 1 | 1kb ladder |
| 2 | Pλ-RBS-luxI-DT(PCR product) |
| 3 | Pbad/araC-RBS-RFP(PCR product) |
Colony PCR
| Sample | base pair |
|---|---|
| 9/11 apt12-1M(pSB1C3) 1 | 513 |
| 9/11 apt12-1M(pSB1C3) 2 | 513 |
| 9/11 Fusion3m2a09ttenuator(pSB1C3) 1 | 609 |
| 9/11 Fusion6 antisense(pSB1C3)1 | 431 |
| 9/11 aptamer 12-P(pSB1C3)1 | 515 |
| 9/11 Fusion1 antisense(pSB1C3) 1 | 420 |
| 9/11 Fusion1 antisense(pSB1C3) 2 | 420 |
| 9/11 Plac(BBa-R0011) 1 | 293 |
| 9/11 Plac(BBa-R0011) 2 | 293 |
| 9/11 Plac(BBa-R0011) 3 | 293 |
| 9/11 Plac(BBa-R0011) 4 | 293 |
| 9/11 Pcon-luxR-Plux-GFP 1 | 2138 |
| 9/11 Pcon-RBS-lacZα-DT(pSB4K5) 1 | 712 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 5min | 30s | 30s | 2min12s | 30cycles |
Restriction Enzyme Digestion
| 9/5 Pcon | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 1cuts(PstI) | 5.7 | 0µL | 0µL | 0µL | 1µL | 3µL | 3µL | 17.3µL | 30µL |
| NC(PstI) | 1.9 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 6.1µL | 10µL |
| 8/17 DT | EcoRI | Buffer | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|
| 1cut(EcoRI) | 10.6 | 1µL | 3µL | 3µL | 12.4µL | 30µL |
| NC(EcoRI) | 3.5 | 0µL | 1µL | 1µL | 4.5µL | 10µL |
Colony PCR
| Sample | base pair |
|---|---|
| 9/11 Pcon-pT181attenuator-RBS-lacZα-DT | 965 |
| 9/11 Plux-PBS-lysis1-DT 1 | 613 |
| 9/11 Pcon-pT181attenuator-aptamer12-1R-DT 1 | 859 |
| 9/11 Pcon-pT181attenuator-aptamer12-1R-DT 2 | 859 |
| 9/11 Plux-RBS-lysis3-DT 1 | 1210 |
| 9/11 Plux-RBS-lysis3-DT 2 | 1210 |
| Pcon-Spinach-DT(pSB4K5) 1 | 605 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 5min | 30s | 30s | 1min12s | 30cycles |
Transformation
| Name | Sample | Competent Cells | Plate |
|---|---|---|---|
| 9/7 pSB4K5 | 2µL | 20µL | Amp |
Electrophoresis
1
| Lane | Sample |
|---|---|
| 1 | Fusion6 antisense |
| 2 | Fusion1 antisense-1 |
| 3 | Fusion1 antisense-2 |
| 4 | 100bp ladder |
| 5 | Plac(BBa-R0011) 1 |
| 6 | Plac(BBa-R0011) 2 |
| 7 | Plac(BBa-R0011) 3 |
| 8 | Plac(BBa-R0011) 4 |
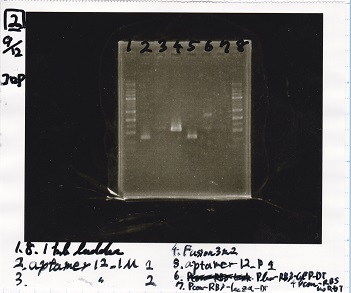
2
| Lane | Sample |
|---|---|
| 1 | 1kb ladder |
| 2 | aptamer12-1M 1 |
| 3 | aptamer12-1M 2 |
| 4 | Fusion3m2 attenuator |
| 5 | aptamer12-P1 |
| 6 | Plux-RBS-GFP-DT-Pcon-RBS-luxR-DT |
| 7 | Pcon-RBS-lacZα=DT |
| 8 | 1kb ladder |
Electrophoresis
| Lane | Sample | Enzyme |
|---|---|---|
| 1 | Pcon-pT181attenuator-RBS-lacZα-DT 1 | -- |
| 2 | Pcon-pT181attenuator-RBS-lacZα-DT 2 | -- |
| 3 | Plux-RBS-lysis1-DT 1 | -- |
| 4 | Pcon-pT181attenuator-aptamer12-1R-DT 1 | -- |
| 5 | Pcon-pT181attenuator-aptamer12-1R-DT 2 | -- |
| 6 | Plux-RBS-lysis3-DT 1 | -- |
| 7 | 1kbp ladder | -- |
| 8 | Plux-RBS-lysis3-DT 2 | -- |
| 9 | Pcon-spinach-DT(pSB4K5) 1 | -- |
| 10 | 9/5 Pcon | PstI |
| 11 | 9/5 Pcon NC | -- |
| 12 | 8/17 DT | EcoRI |
| 13 | 8/17 DT | -- |
Liquid Culture
| Sample | medium |
|---|---|
| 9/11Fusion6 antisense-1 | Plusgrow medium(+CP) |
| Plac(BBa-R0011)-3 | Plusgrow medium(+Amp) |
| Plac(BBa-R0011)-4 | Plusgrow medium(+Amp) |
| Fusion3m2 attenuator -1 | Plusgrow medium(+CP) |
| Pλ-luxI-2 | Plusgrow medium(+Amp) |
| 9/11 aptamer12-1M-2 | Plusgrow medium(+CP) |
Colony PCR
| Sample | base pair |
|---|---|
| 9/11 Fusion1 antisense 3 | 394 |
| 9/11 Fusion1 antisense 4 | 394 |
| 9/11 aptamer12-P 2 | 378 |
| 9/11 aptamer12-P 3 | 378 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 5min | 30s | 30s | 36s | 30cycles |
Electrophoresis
| Lane | Sample | Enzyme |
|---|---|---|
| 1 | Fusion1 antisense 2 | -- |
| 2 | Fusion1 antisense 3 | -- |
| 3 | 1kb ladder | -- |
| 4 | apt12-P 2 | -- |
| 5 | apt12-P 3 | -- |
Restriction Enzyme Digestion
| 9/12 RBS-lysis2-DT | XbaI | PstI | Buffer | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 5.2 | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 16.8µL | 30µL |
| NC | 0.3µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.7µ | 10µL |
| 9/12 RBS-lysis3-DT | XbaI | PstI | Buffer | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 5.2 | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 16.8µL | 30µL |
| NC | 0.3µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.7µ | 10µL |
| 9/11 RBS-lysis1-DT | XbaI | PstI | Buffer | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 8.5 | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 13.5µL | 30µL |
| NC | 0.4µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.6µ | 10µL |
| 8/20 J23100-RBS-luxR -DT | EcoRI | Buffer | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|
| 1cut | 5.8 | 1.0µL | 3.0µL | 3.0µL | 17.2µL | 30µL |
| NC | 0.3µL | 0µL | 1.0µL | 1.0µL | 7.7µ | 10µL |
| 9/10 pSB1C3 | EcoRI | SpeI | Buffer | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2cuts | 8.1 | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 13.9µL | 30µL |
| NC | 0.4µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.6µ | 10µL |
| 9/12 Plux | PstI | Buffer | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|
| 1cut | 12.0 | 1.0µL | 3.0µL | 3.0µL | 11.0micro;L | 30µL |
| NC | 0.6µL | 0µL | 1.0µL | 1.0µL | 7.4µ | 10µL |
| 9/10 pSB1C3 | XbaI | PstI | Buffer | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2cuts | 8.1 | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 13.9µL | 30µL |
Electrophoresis
| Lane | Sample | Enzyme1 | Enzymel2 |
|---|---|---|---|
| 1 | 9/12 RBS-lysis2-DT | XbaI | PstI |
| 2 | 9/12 RBS-lysis2-DT NC | -- | -- |
| 3 | 9/11 RBS-lysis1-DT | XbaI | PstI |
| 4 | 9/11 RBS-lysis1-DT NC | -- | -- |
| 5 | 9/11 RBS-lysis3-DT | XbaI | PstI |
| 6 | 9/11 RBS-lysis3-DT NC1 | -- | -- |
| 7 | 1kb ladder | -- | -- |
| 8 | 100bp ladder | -- | -- |
| 9 | J23100-RBS-luxR-DT | EcoRI | -- |
| 10 | J23100-RBS-luxR-DT | -- | -- |
| 11 | 9/10 pSB1C3 | EcoRI | SpeI |
| 12 | 9/10 pSB1C3 NC | -- | --- |
| 13 | 9/10 pSB1C3 | XbaI | PstI |
| 14 | 9/12 Plux | PstI | -- |
| 15 | 9/12 Plux NC | -- | -- |
Observation through a confocal microscope
We used the liquid culture media with 200μL 9/10Pcon-pT181antisense-spinach-DT& 200μL 9/10 Pcon-spinach-DT& 200μL 9/10 JM109(overnight culture).
We translated them into each 1.5mL tube.
We elminated each supernatant using a 5000xg centrifuge for 1minute,.
Then,we resuspended with 100µL M9(distilled water) and eliminated each supernatant using a 5000xg centrifuge for 1 minute.x2
Adding 100µL M9(in 20µM DFHBI), we resuspended the pret.
15min after incubating in shield light, we eliminated each supernatant using a 5000xg centrifuge for 1 minute.
We looked at the fluorescence of the pret.
We failed to observe it.
Liquid Culture
| Sample | medium |
|---|---|
| 9/10 Pcon-pT181antisense-spinach-DT | Plusgrow medium |
| 9/10 Pcon-spinach-DT | Plusgrow medium |
| 9/1 spinach-DT(RNA master plate) | Plusgrow medium |
 "
"