Team:Tokyo Tech/Experiment/pSB-M13 Plasmid Assay
From 2013.igem.org
Shinya0825 (Talk | contribs) |
|||
| (43 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{tokyotechmenudark}} | {{tokyotechmenudark}} | ||
| - | |||
| - | |||
<div id="text-area"><br> | <div id="text-area"><br> | ||
| + | <div class="box" id="title"> | ||
| + | <p style="line-height:0em; text-indent:0em;" name="top">pSB-M13 Plasmid Assay</p> | ||
| + | </div> | ||
<div class="box"> | <div class="box"> | ||
| - | + | <h1>1. Introduction</h1> | |
| - | + | ||
| - | </ | + | |
<h2> | <h2> | ||
| - | <p> | + | <p>We confirmed that M13 phage genes and M13 origin worked properly with pSB origin, using a plaque forming assay. The plasmid construction is as follows. First, from M13mp18 phage vector, we amplified the sequence that includes from <i>g2p</i> RBS to M13 origin (with prefix and suffix sequence), by PCR (Fig. 3-3-1). Second, we inserted the amplified sequence to pSB3K3 backbone. Finally, upstream of the RBS-<i>g2p</i>, we inserted PlacI<sup>q</sup> (constitutive) promoter([https://2013.igem.org/Team:Tokyo_Tech/Project/M13_supplement#1._M13_plasmid_construction See more about our plasmid construction]). |
</p> | </p> | ||
| + | [[Image:Titech2013_M13_PlacIq_Fig.1.PNG|600px|thumb|center|Fig. 3-3-1. Our designed plasmid constrction]] | ||
| + | |||
</h2> | </h2> | ||
| - | < | + | <h1>2. Materials and Methods</h1> |
| - | </ | + | <h3>2-1. Plasmid construction</h3> |
| - | + | ||
| - | + | ||
| - | <h3>2. | + | |
| - | </h3> | + | |
<h2> | <h2> | ||
| - | + | <p>pSB3K3-PlacI<sup>q</sup>-M13-Plac-<i>GFP</i> ([http://parts.igem.org/Part:BBa_K1139020 BBa_K1139020]) | |
| - | <p>pSB3K3- | + | |
</p> | </p> | ||
| - | <p> | + | <p>pSB3K3-M13-Plac-<i>GFP</i> ([http://parts.igem.org/Part:BBa_K1139022 BBa_K1139022]) |
| + | </p></h2> | ||
| + | <h3>2-2. Strains</h3> | ||
| + | <h2> | ||
| + | <p>DH5alpha (<i>E. coli</i> of high competence) | ||
</p> | </p> | ||
| - | + | <p>JM109 (F+ strain <i>E. coli</i>) | |
| - | + | </p></h2> | |
| - | <p> | + | <h3>2-3. Media</h3> |
| - | </p> | + | <h2> |
| - | < | + | <div style="margin-left:80px;"> |
| - | < | + | LB |
| - | + | {| class="wikitable" cellpadding="6" | |
| - | + | |Bacto tryptone||10 g/L | |
| - | + | |- | |
| - | + | |Yeast Extract|| 5 g/L | |
| - | + | |- | |
| - | + | |NaCl||10 g/L | |
| - | + | |} | |
| - | + | ||
| - | + | YT plate | |
| - | + | {| class="wikitable" cellpadding="6" | |
| - | + | |Bacto tryptone|| 8 g/L | |
| - | + | |- | |
| - | + | |Yeast Extract|| 5 g/L | |
| - | + | |- | |
| - | + | |NaCl|| 5 g/L | |
| - | + | |- | |
| - | + | |Agarose||15 g/L | |
| - | + | |} | |
| - | + | ||
| - | + | YT soft agar | |
| - | + | {| class="wikitable" cellpadding="6" | |
| - | + | |Bacto tryptone||8 g/L | |
| - | + | |- | |
| - | + | |Yeast Extract||5 g/L | |
| - | + | |- | |
| - | + | |NaCl||5 g/L | |
| - | + | |- | |
| - | + | |Agarose||6 g/L | |
| - | + | |} | |
| - | + | </div> | |
| - | + | ||
| - | </ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</h2> | </h2> | ||
| - | <h3> | + | <h3>2-4. Protocol</h3> |
| - | </h3> | + | |
<h2> | <h2> | ||
| - | < | + | <ol> |
| - | </ | + | <em align="center">Preparation</em> |
| - | < | + | <li>Transform DH5alpha with pSB3K3-PlacI<sup>q</sup>-M13. |
| - | < | + | <li>Grow overnight culture of the transformed DH5alpha and JM109 at 37°C.<br> |
| - | + | <em align="center">Plaque formation</em> | |
| - | < | + | <li>Spin the overnight culture of the transformed DH5alpha at 9,000g for 1 min. |
| - | </ | + | <li>Pipette the supernatant into a 1.5 mL tube. |
| - | + | <li>Dilute it 100 times with water. (-> phage-particle-solution) | |
| - | < | + | <li>Melt YT soft agar using a microwave. |
| - | </ | + | <li>Dispense 3.5 mL of the melted soft agar to a new round tube, and keep them at 50°C in an incubator. |
| - | + | <li>Dispense 400 µL of overnight culture of JM109 to a 1.5 mL tube. | |
| - | <p> | + | <li>Into the 1.5 mL tube, add 100 µL of the phage-particle-solution. |
| + | <li>Transfer all the content of the 1.5 mL tube to the dispensed soft agar, mix them, and decant all of it on a YT plate. | ||
| + | </ol></h2> | ||
| + | <h1>3. Result</h1> | ||
| + | <h2> | ||
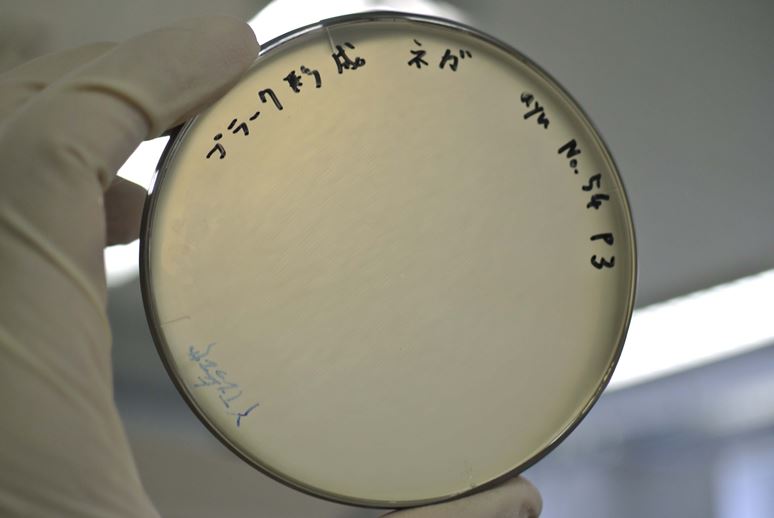
| + | <p>The result of the plaque forming assay is showed in Fig. 3-3-2, 3-3-3. M13 phage genes and M13 origin worked together with <i>lacI<sup>q</sup></i> promoter and pSB origin. | ||
</p> | </p> | ||
| + | [[Image:Titech2013_M13_PlacIq_A.jpg|600px|thumb|center|Fig. 3-3-2. The plaques were formed using JM109 overnight culture and phage-particle-solution. The phage particles were obtained from the supernatant of the overnight culture of transformed DH5alpha (with pSB3K3- PlacI<sup>q</sup>-M13) at 9,000g. A mixture of 3.5 mL YT soft agar, 100 µL of 100X supernatant and 400 µL of overnight culture of JM109 was poured on a YT plate.]] | ||
| + | |||
| + | [[Image:Titech2013_M13_PlacIq_B.jpg|600px|thumb|center|Fig. 3-3-3. A mixture of only 3.5 mL YT soft agar and 500 µL of overnight culture of JM109 was poured on a YT plate.]] | ||
</h2> | </h2> | ||
| - | |||
| - | |||
| - | |||
| - | |||
</div><br> | </div><br> | ||
| + | <html><div align="center"><a href="https://2013.igem.org/Team:Tokyo_Tech/Experiment/pSB-M13_Plasmid_Assay#top"><img src="https://static.igem.org/mediawiki/2013/f/f0/Titeh2013_backtotop.png" width="200px"></a></div></html> | ||
</div> | </div> | ||
| - | |||
| - | |||
Latest revision as of 17:14, 28 October 2013
pSB-M13 Plasmid Assay
Contents |
1. Introduction
We confirmed that M13 phage genes and M13 origin worked properly with pSB origin, using a plaque forming assay. The plasmid construction is as follows. First, from M13mp18 phage vector, we amplified the sequence that includes from g2p RBS to M13 origin (with prefix and suffix sequence), by PCR (Fig. 3-3-1). Second, we inserted the amplified sequence to pSB3K3 backbone. Finally, upstream of the RBS-g2p, we inserted PlacIq (constitutive) promoter(See more about our plasmid construction).
2. Materials and Methods
2-1. Plasmid construction
pSB3K3-PlacIq-M13-Plac-GFP ([http://parts.igem.org/Part:BBa_K1139020 BBa_K1139020])
pSB3K3-M13-Plac-GFP ([http://parts.igem.org/Part:BBa_K1139022 BBa_K1139022])
2-2. Strains
DH5alpha (E. coli of high competence)
JM109 (F+ strain E. coli)
2-3. Media
LB
Bacto tryptone 10 g/L
Yeast Extract 5 g/L
NaCl 10 g/L
YT plate
Bacto tryptone 8 g/L
Yeast Extract 5 g/L
NaCl 5 g/L
Agarose 15 g/L
YT soft agar
Bacto tryptone 8 g/L
Yeast Extract 5 g/L
NaCl 5 g/L
Agarose 6 g/L
LB
| Bacto tryptone | 10 g/L |
| Yeast Extract | 5 g/L |
| NaCl | 10 g/L |
YT plate
| Bacto tryptone | 8 g/L |
| Yeast Extract | 5 g/L |
| NaCl | 5 g/L |
| Agarose | 15 g/L |
YT soft agar
| Bacto tryptone | 8 g/L |
| Yeast Extract | 5 g/L |
| NaCl | 5 g/L |
| Agarose | 6 g/L |
2-4. Protocol
Preparation
- Transform DH5alpha with pSB3K3-PlacIq-M13.
- Grow overnight culture of the transformed DH5alpha and JM109 at 37°C.
Plaque formation
- Spin the overnight culture of the transformed DH5alpha at 9,000g for 1 min.
- Pipette the supernatant into a 1.5 mL tube.
- Dilute it 100 times with water. (-> phage-particle-solution)
- Melt YT soft agar using a microwave.
- Dispense 3.5 mL of the melted soft agar to a new round tube, and keep them at 50°C in an incubator.
- Dispense 400 µL of overnight culture of JM109 to a 1.5 mL tube.
- Into the 1.5 mL tube, add 100 µL of the phage-particle-solution.
- Transfer all the content of the 1.5 mL tube to the dispensed soft agar, mix them, and decant all of it on a YT plate.
Plaque formation
3. Result
The result of the plaque forming assay is showed in Fig. 3-3-2, 3-3-3. M13 phage genes and M13 origin worked together with lacIq promoter and pSB origin.

 "
"