Team:Gaston Day School/Project
From 2013.igem.org
| (8 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Team:Gaston_Day_School/Templates/top}} | {{Template:Team:Gaston_Day_School/Templates/top}} | ||
| - | |||
| - | |||
<html> | <html> | ||
<br> | <br> | ||
| Line 8: | Line 6: | ||
{|align="justify" | {|align="justify" | ||
| | | | ||
| - | Serious health problems arise throughout the world due to the common water contaminants, heavy metals. The Agency for Toxic Substances and Disease | + | Serious health problems arise throughout the world due to the common water contaminants, heavy metals. The Agency for Toxic Substances and Disease Registry (ASTDR) released a Priority List of Hazardous Substances of which heavy metals accounted for almost half of the top 10 substances. These substances can ultimately cause cancer along with other symptoms in affected systems like the lungs, blood, digestive system, and nervous system. Therefore, our 2012 team constructed a set of sensors that detect heavy metal contaminants in water. Among our Arsenic, Lead and Cadmium Sensors, we decided to improve the sensitivity of the Cadmium Sensor to provide more accurate and medically relevant levels of detection. This sensor previously paired a promoter responsive to cadmium with GFP as a reporter. To improve this sensor, it was combined with an activator and a stronger promoter to amplify the signal as it reached the GFP reporter. Use of the detector could potentially save lives around the world through earlier detection of the contamination. |
| - | + | One could also create a kit that would include all necessary components for running tests and then decontaminating the resulting growth to prevent release of the engineered bacteria into the environment. It should also include a very simple mechanism for killing or denaturing the bacteria in the detector kit – bleach. Bleach is highly effective at killing bacteria and is readily available to the average person. | |
|- | |- | ||
| | | | ||
| Line 19: | Line 17: | ||
</html> | </html> | ||
<div style="text-align: left;"> | <div style="text-align: left;"> | ||
| - | We | + | We originally decided to approach the sensitivity problem two ways. First we used the 2011 UC Davis iGEM team's [https://2011.igem.org/Team:UC_Davis/Protocols#ER-PCR mutagenic PCR protocol] to create mutations in the DNA of the entire Cadmium Sensor and just the cadmium promoter. After multiple runs, we hoped to find at least one colony that had increased sensitivity due to these mutations. |
| - | Step 1: We digested the promoter responsive to | + | Second we utilized the 2007 Cambridge iGEM team's [https://2007.igem.org/wiki/index.php/Cambridge/Amplifier_project sensitivity tuners] and began building the improved Cadmium Sensor using NEB's BioBrick Construction Kit and creating intermediate parts to form the final construct. |
| + | |||
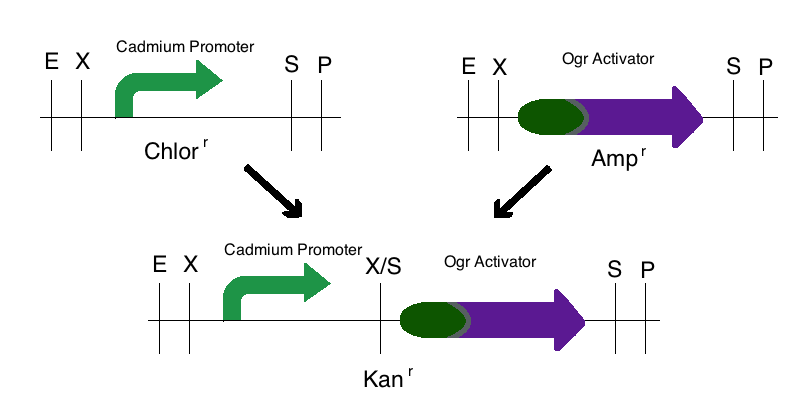
| + | Step 1: We digested the promoter responsive to cadmium (BBa_K174015) with EcoRI and SpeI and digested an activator (either ogr activator, BBa_I746350, or the delta activator, BBa_I746352) with XbaI and PstI. These parts were then ligated to a Kanamycin vector that was digested with EcoRI and PstI. This created two intermediate parts that were selectively grown on Kanamycin plates. | ||
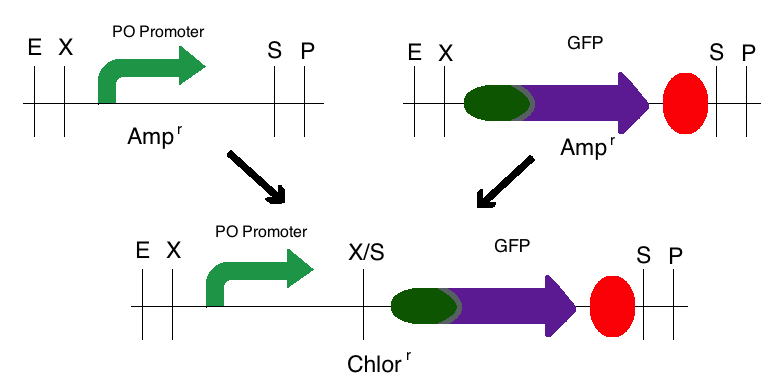
Step 2: The same process was used for the second intermediate part in which the PO Promoter (BBa_I746361) was the upstream part and the GFP plasmid (BBa_I13401) was the downstream part. However, these digested fragments were transferred to a Chloramphenicol vector. This produced one intermediate that was selectively grown on a Chloramphenicol plate. | Step 2: The same process was used for the second intermediate part in which the PO Promoter (BBa_I746361) was the upstream part and the GFP plasmid (BBa_I13401) was the downstream part. However, these digested fragments were transferred to a Chloramphenicol vector. This produced one intermediate that was selectively grown on a Chloramphenicol plate. | ||
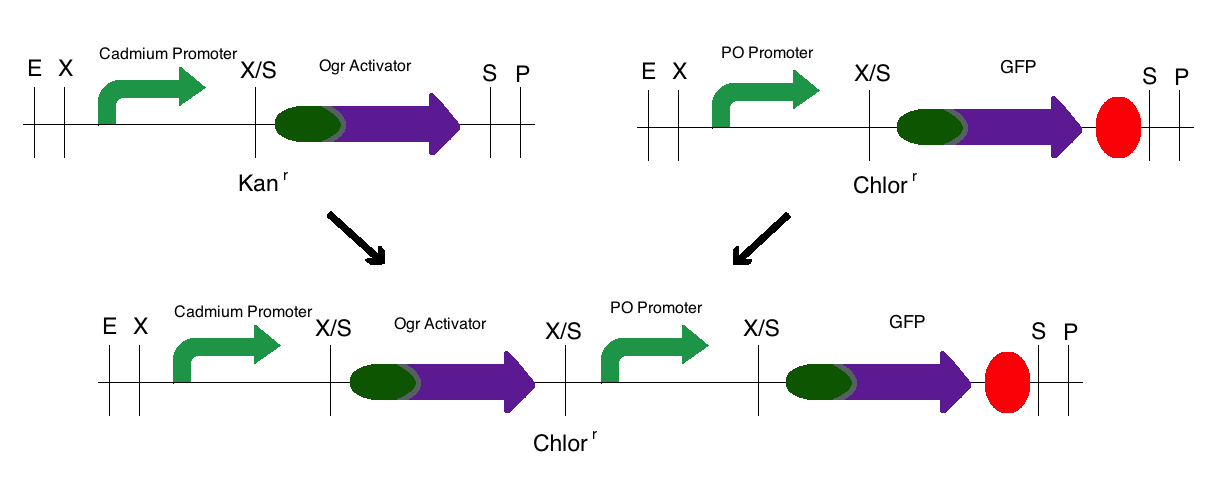
| - | Step 3: The final construct was made in the same process where the intermediates of step 1 became upstream parts and the intermediate of step 2 became the downstream part. These parts were then mixed and ligated to the provided, linearized pSB1C3 plasmid. The resulting colonies were tested for responsive GFP production from the addition of | + | Step 3: The final construct was made in the same process where the intermediates of step 1 became upstream parts and the intermediate of step 2 became the downstream part. These parts were then mixed and ligated to the provided, linearized pSB1C3 plasmid. The resulting colonies were tested for responsive GFP production from the addition of cadmium. Modeling of the process is shown below. |
| - | ''Note:'' The same methods were used for constructing a sensor with a delta activator instead of an ogr activator. The images below also | + | ''Note:'' The same methods were used for constructing a sensor with a delta activator instead of an ogr activator. The images below also models that part. |
| - | <div class="center" style="width:auto; margin-left:auto; margin-right:auto;">[[Image:Intermediate_1.png| | + | <div class="center" style="width:auto; margin-left:auto; margin-right:auto;">[[Image:Intermediate_1.png|750px]] |
</div> | </div> | ||
| - | <div class="center" style="width:auto; margin-left:auto; margin-right:auto;">[[Image:Intermediate_2.png| | + | <div class="center" style="width:auto; margin-left:auto; margin-right:auto;">[[Image:Intermediate_2.png|750px]] |
</div> | </div> | ||
| Line 44: | Line 44: | ||
</html> | </html> | ||
<div style="text-align: left;"> | <div style="text-align: left;"> | ||
| - | We tested out detectors (protocol can be found [https://static.igem.org/mediawiki/2013/3/3a/Gds13.9-3.FINAL.pdf here]) and measured the sensitivity using a spectrophotometer. We measured in | + | We tested out detectors (protocol can be found [https://static.igem.org/mediawiki/2013/3/3a/Gds13.9-3.FINAL.pdf here]) and measured the sensitivity using a spectrophotometer. We measured in Fluor./OD which is the amount of fluorescence per OD (at 600nm). At first, we encountered many problems. We noticed that all of our dilutions, including the control were approximately the same. We realized that LB (Luria Broth, our growth medium) fluoresces naturally; hence, it masked the effects of the GFP. To solve this problem, we grew our bacteria in LB, centrifuged it, then resuspended in PBS (phosphate buffer solution). Lastly, our spectrophotometer still continued to show the same fluorescence reading for most samples except the control. We then realized that the readings had maxed. Serial dilutions were used to determine accurate measurements. |
<html> | <html> | ||
| Line 51: | Line 51: | ||
</html> | </html> | ||
<div style="text-align: left;"> | <div style="text-align: left;"> | ||
| + | The mutagenesis of the Cadmium Sensor and promoter gave us multiple samples, but nearly all of the samples had either the same level of sensitivity or lower. Those that showed increased sensitivity in preliminary tests revealed only slightly increased GFP output that was not stable. | ||
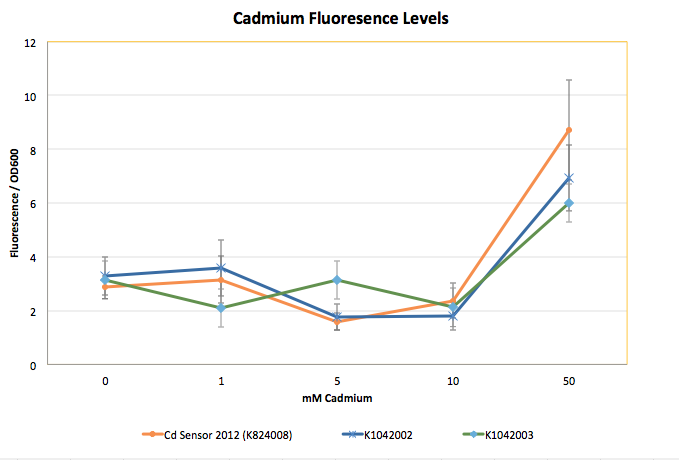
The graph shown below represents our results (in Fluorescence per OD600) for the detector with the delta activator (K1042002) and the detector with the ogr activator (K1042003). | The graph shown below represents our results (in Fluorescence per OD600) for the detector with the delta activator (K1042002) and the detector with the ogr activator (K1042003). | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| + | [[File:Cadmium_Fluoresence_Levels.png|center|650px|Not much improvement.]] | ||
<br/> | <br/> | ||
| - | + | ||
| - | + | Again we discovered only slightly increased output with the addition of the sensitivity tuners. These results, coupled with a size check of our final parts being much smaller than expected, led us to believe that we had not actually created the final construct we designed. This error could be attributed to the fact that our second intermediate part was already in a chloramphenicol backbone and so trying to do a ligation from one chloramphenicol backbone to another would not guarantee us the part we wanted. After doing another size check of the intermediates, we were able to submit the PO Promoter and GFP combination (K10420013), and we are now in the process of constructing and carrying out preliminary testing on our improved Cadmium Sensor. | |
Latest revision as of 20:28, 27 September 2013
Overall Project
|
Serious health problems arise throughout the world due to the common water contaminants, heavy metals. The Agency for Toxic Substances and Disease Registry (ASTDR) released a Priority List of Hazardous Substances of which heavy metals accounted for almost half of the top 10 substances. These substances can ultimately cause cancer along with other symptoms in affected systems like the lungs, blood, digestive system, and nervous system. Therefore, our 2012 team constructed a set of sensors that detect heavy metal contaminants in water. Among our Arsenic, Lead and Cadmium Sensors, we decided to improve the sensitivity of the Cadmium Sensor to provide more accurate and medically relevant levels of detection. This sensor previously paired a promoter responsive to cadmium with GFP as a reporter. To improve this sensor, it was combined with an activator and a stronger promoter to amplify the signal as it reached the GFP reporter. Use of the detector could potentially save lives around the world through earlier detection of the contamination. One could also create a kit that would include all necessary components for running tests and then decontaminating the resulting growth to prevent release of the engineered bacteria into the environment. It should also include a very simple mechanism for killing or denaturing the bacteria in the detector kit – bleach. Bleach is highly effective at killing bacteria and is readily available to the average person. |
Project Details
We originally decided to approach the sensitivity problem two ways. First we used the 2011 UC Davis iGEM team's mutagenic PCR protocol to create mutations in the DNA of the entire Cadmium Sensor and just the cadmium promoter. After multiple runs, we hoped to find at least one colony that had increased sensitivity due to these mutations.
Second we utilized the 2007 Cambridge iGEM team's sensitivity tuners and began building the improved Cadmium Sensor using NEB's BioBrick Construction Kit and creating intermediate parts to form the final construct.
Step 1: We digested the promoter responsive to cadmium (BBa_K174015) with EcoRI and SpeI and digested an activator (either ogr activator, BBa_I746350, or the delta activator, BBa_I746352) with XbaI and PstI. These parts were then ligated to a Kanamycin vector that was digested with EcoRI and PstI. This created two intermediate parts that were selectively grown on Kanamycin plates.
Step 2: The same process was used for the second intermediate part in which the PO Promoter (BBa_I746361) was the upstream part and the GFP plasmid (BBa_I13401) was the downstream part. However, these digested fragments were transferred to a Chloramphenicol vector. This produced one intermediate that was selectively grown on a Chloramphenicol plate.
Step 3: The final construct was made in the same process where the intermediates of step 1 became upstream parts and the intermediate of step 2 became the downstream part. These parts were then mixed and ligated to the provided, linearized pSB1C3 plasmid. The resulting colonies were tested for responsive GFP production from the addition of cadmium. Modeling of the process is shown below.
Note: The same methods were used for constructing a sensor with a delta activator instead of an ogr activator. The images below also models that part.
The Experiments
We tested out detectors (protocol can be found here) and measured the sensitivity using a spectrophotometer. We measured in Fluor./OD which is the amount of fluorescence per OD (at 600nm). At first, we encountered many problems. We noticed that all of our dilutions, including the control were approximately the same. We realized that LB (Luria Broth, our growth medium) fluoresces naturally; hence, it masked the effects of the GFP. To solve this problem, we grew our bacteria in LB, centrifuged it, then resuspended in PBS (phosphate buffer solution). Lastly, our spectrophotometer still continued to show the same fluorescence reading for most samples except the control. We then realized that the readings had maxed. Serial dilutions were used to determine accurate measurements.
Results
The mutagenesis of the Cadmium Sensor and promoter gave us multiple samples, but nearly all of the samples had either the same level of sensitivity or lower. Those that showed increased sensitivity in preliminary tests revealed only slightly increased GFP output that was not stable.
The graph shown below represents our results (in Fluorescence per OD600) for the detector with the delta activator (K1042002) and the detector with the ogr activator (K1042003).
 "
"