Template:Kyoto/Notebook/Sep 22
From 2013.igem.org
(Difference between revisions)
(→Transformation) |
(→PCR) |
||
| (29 intermediate revisions not shown) | |||
| Line 15: | Line 15: | ||
|} | |} | ||
</div> | </div> | ||
| + | [[File:Igku_0903_E1.jpg]]<br> | ||
| + | [[File:Igku_0922_E2.jpg]]<br> | ||
| + | |||
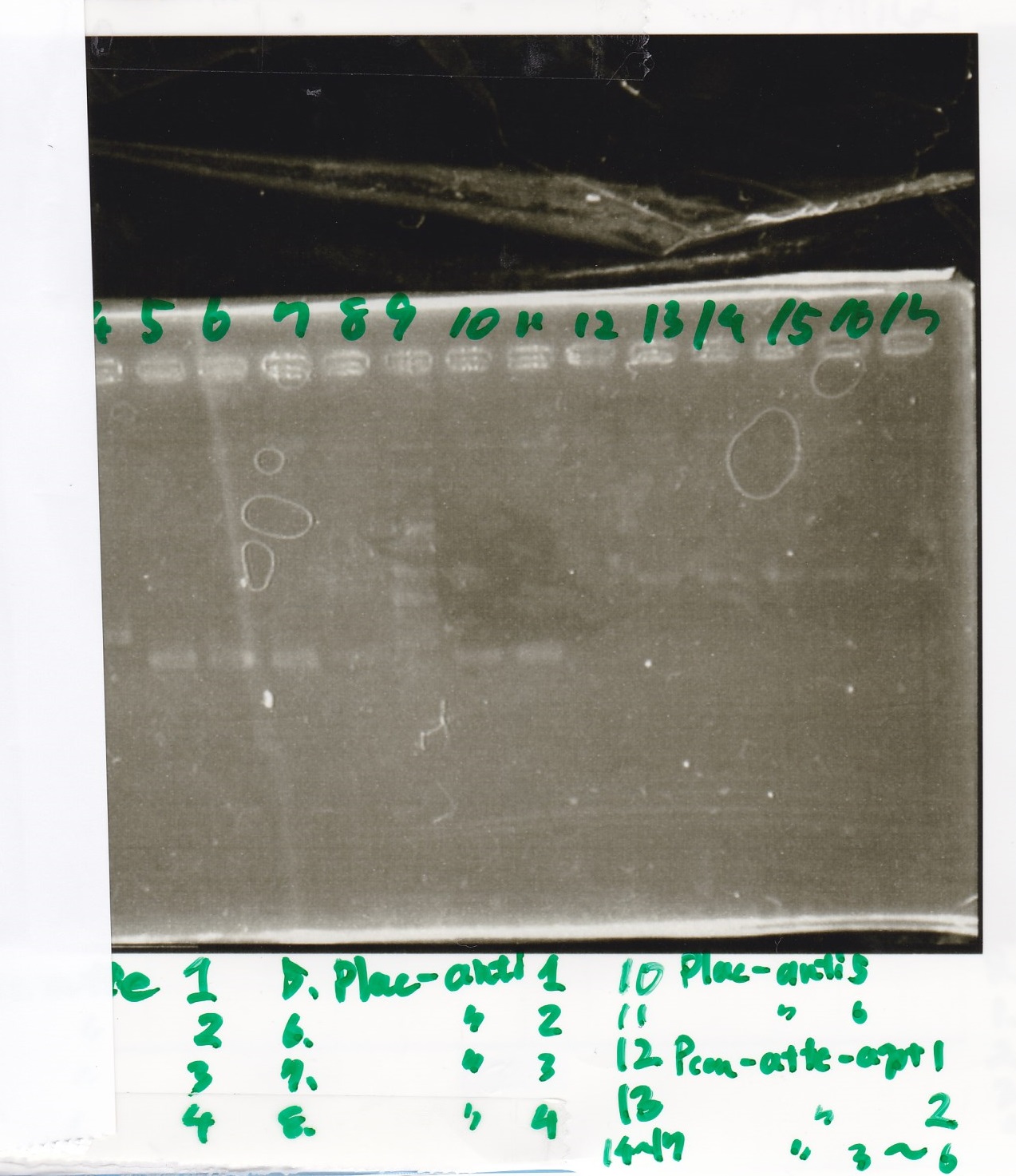
| + | ===Electrophoresis=== | ||
| + | <div class="experiment"> | ||
| + | <span class="author">Nakamoto</span> | ||
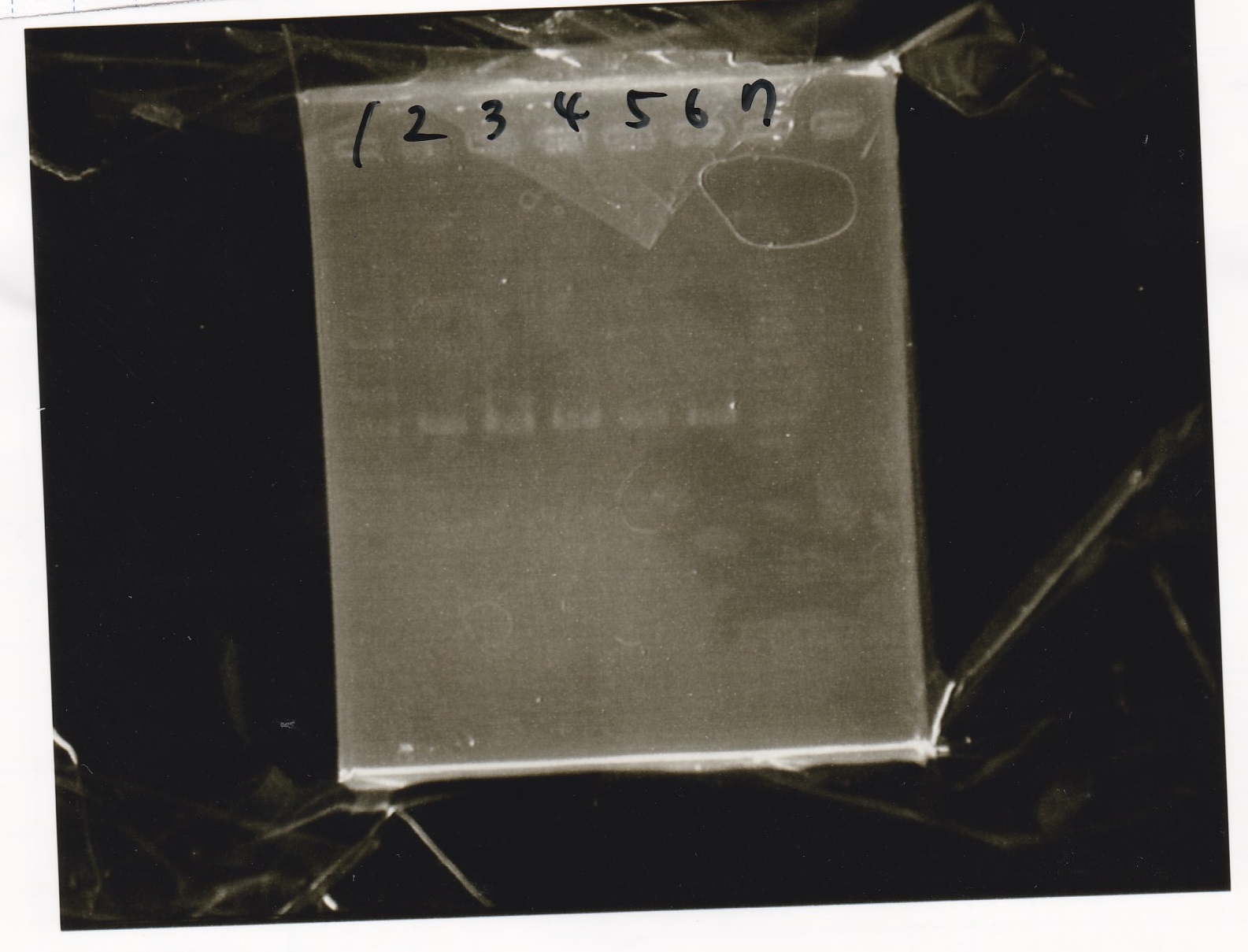
| + | {| class="wikitable" | ||
| + | !Lane||Sample | ||
| + | |- | ||
| + | |1||Pcon-apt12_1R-DT(E-1A) | ||
| + | |- | ||
| + | |2||Pcon-spinach-DT(E-1A) | ||
| + | |- | ||
| + | |3||Pcon-pT181 attenuator(E-1A) | ||
| + | |- | ||
| + | |4||DT(1-3) | ||
| + | |- | ||
| + | |5||tetR aptamer 12_1R-DT(1-3) | ||
| + | |- | ||
| + | |6||100bp ladder | ||
| + | |- | ||
| + | |7||Pcon-pT181 attenuator(1-SA) | ||
| + | |- | ||
| + | |8||spinach-DT(2-1) | ||
| + | |- | ||
| + | |9||Pcon-pT181 attenuator(E-0) | ||
| + | |- | ||
| + | |10||Pcon-pT181 attenuator(2-SA) | ||
| + | |- | ||
| + | |11||spinach-DT(3-2) | ||
| + | |- | ||
| + | |12||Ptet-PT181 anisense | ||
| + | |} | ||
| + | [[File:Igku_0922_E3.jpg]]<br> | ||
| + | [[File:Igku_0922_E4.jpg]]<br> | ||
===Miniprep=== | ===Miniprep=== | ||
| Line 51: | Line 86: | ||
|5min||30s||30s||2min10s||30cycles | |5min||30s||30s||2min10s||30cycles | ||
|} | |} | ||
| + | [[File:igku_0922_E5.jpg]]<br> | ||
| + | [[File:igku_0922_E6.jpg]]<br> | ||
===Gel Extraction=== | ===Gel Extraction=== | ||
| Line 95: | Line 132: | ||
|} | |} | ||
</div> | </div> | ||
| + | [[File:Igku 0922 G1.jpg]]<br> | ||
| + | [[File:Igku 0922 G2.jpg]]<br> | ||
| + | [[File:Igku 0922 G3.jpg]]<br> | ||
| + | [[File:Igku 0922 G4.jpg]]<br> | ||
| - | === | + | ===Restriction Enzyme Digestion=== |
<div class="experiment"> | <div class="experiment"> | ||
<span class="author">No name</span> | <span class="author">No name</span> | ||
| Line 113: | Line 154: | ||
|2cuts||16µL||0µL||1µL||1µL||3µL||3µL||14µL||30µL | |2cuts||16µL||0µL||1µL||1µL||3µL||3µL||14µL||30µL | ||
|- | |- | ||
| - | |NC||0.8µL||0µL | + | |NC||0.8µL||0µL||0µL||0µL||1µL||1µL||7.2µL||10µL |
|} | |} | ||
| Line 124: | Line 165: | ||
|- | |- | ||
|} | |} | ||
| + | [[File:igku_0922_E7.jpg]]<br> | ||
===Gel Extraction=== | ===Gel Extraction=== | ||
| Line 171: | Line 213: | ||
|Ptet-antisense PT181(1-3)||16.8||1.74||0.78 | |Ptet-antisense PT181(1-3)||16.8||1.74||0.78 | ||
|} | |} | ||
| + | [[File:Igku 0922 G5.jpg]]<br> | ||
| + | [[File:Igku 0922 G6.jpg]]<br> | ||
| + | [[File:Igku 0922 G7.jpg]]<br> | ||
| + | [[File:Igku 0922 G8.jpg]]<br> | ||
| + | [[File:Igku 0922 G9.jpg]]<br> | ||
| + | [[File:Igku 0922 G10.jpg]]<br> | ||
===Colony PCR=== | ===Colony PCR=== | ||
| Line 189: | Line 237: | ||
|PconPT181antisense-spinach-DT(9~15)||723 | |PconPT181antisense-spinach-DT(9~15)||723 | ||
|} | |} | ||
| - | + | [[File:igku_0922_E8.jpg]]<br> | |
===Ligation=== | ===Ligation=== | ||
| Line 197: | Line 245: | ||
!state||colspan="2"|Vector||colspan="2"|Inserter||Ligation High ver.2 | !state||colspan="2"|Vector||colspan="2"|Inserter||Ligation High ver.2 | ||
|- | |- | ||
| - | |experiment||9/13 Pcon-PT181 attenuator (SpeI & PstI)||1.7 µL||9/18 aptamer-1R-DT (XbaI & PstI)||5.4µL|| | + | |experiment||9/13 Pcon-PT181 attenuator (SpeI & PstI)||1.7 µL||9/18 aptamer-1R-DT (XbaI & PstI)||5.4µL|| 3.5µL |
|- | |- | ||
| - | |experiment||9/12 Plac(SpeI & PstI)||2.4 µL||9/18 aptamer-1R-DT (XbaI & PstI)||6.1µL|| | + | |experiment||9/12 Plac(SpeI & PstI)||2.4 µL||9/18 aptamer-1R-DT (XbaI & PstI)||6.1µL||3.5 µL |
|- | |- | ||
| - | |experiment||9/13 Ptet(SpeI & PstI)||1.7 µL||9/10 Plux-RBS-GFP-DT||7.0 µL|| | + | |experiment||9/13 Ptet(SpeI & PstI)||1.7 µL||9/10 Plux-RBS-GFP-DT||7.0 µL||4.4 µL |
|- | |- | ||
| - | |experiment||9/21 RBS-GFP-DT (EcoRI & XbaI)||0.6 µL||9/18 aptamer 12-1R-DT (XbaI & PstI)||5.6µL|| | + | |experiment||9/21 RBS-GFP-DT (EcoRI & XbaI)||0.6 µL||9/18 aptamer 12-1R-DT (XbaI & PstI)||5.6µL||3.1 µL |
|- | |- | ||
| - | |experiment||9/13 PSB1C3(XbaI & PstI)||1.9 µL||9/16 PT181 attenuator(XbaI & PstI)||4.6µL|| | + | |experiment||9/13 PSB1C3(XbaI & PstI)||1.9 µL||9/16 PT181 attenuator(XbaI & PstI)||4.6µL||3.3 µL |
|- | |- | ||
|} | |} | ||
| Line 241: | Line 289: | ||
! ||9/7 Pcon-RBS-tetR-DT||EcoRI||XbaI||SpeI||PstI||BufferD||BSA||MilliQ||total | ! ||9/7 Pcon-RBS-tetR-DT||EcoRI||XbaI||SpeI||PstI||BufferD||BSA||MilliQ||total | ||
|- | |- | ||
| - | |2cut||14µL||1µL||1µL||0µL||0µL||3µL||18.2µL||30µL | + | |2cut||14µL||1µL||1µL||0µL||0µL||3µL||3µL||18.2µL||30µL |
|- | |- | ||
|NC||0.7µL||0µL||0µL||0µL||0µL||1µL||1µL||7.3µL||10µL | |NC||0.7µL||0µL||0µL||0µL||0µL||1µL||1µL||7.3µL||10µL | ||
| Line 263: | Line 311: | ||
|} | |} | ||
incubate 37°C 1hour</div> | incubate 37°C 1hour</div> | ||
| + | [[File:Igku 0922 E9.jpg]]<br> | ||
===Gel Extraction=== | ===Gel Extraction=== | ||
| Line 275: | Line 324: | ||
|} | |} | ||
</div> | </div> | ||
| + | [[File:Igku 0922 G11.jpg]]<br> | ||
| + | [[File:Igku 0922 G12.jpg]]<br> | ||
===Transformation=== | ===Transformation=== | ||
<div class="experiment"> | <div class="experiment"> | ||
| - | <span class="author"> | + | <span class="author">No name</span> |
{| class="wikitable" | {| class="wikitable" | ||
!Name||Sample||Competent Cells||Plate | !Name||Sample||Competent Cells||Plate | ||
| Line 306: | Line 357: | ||
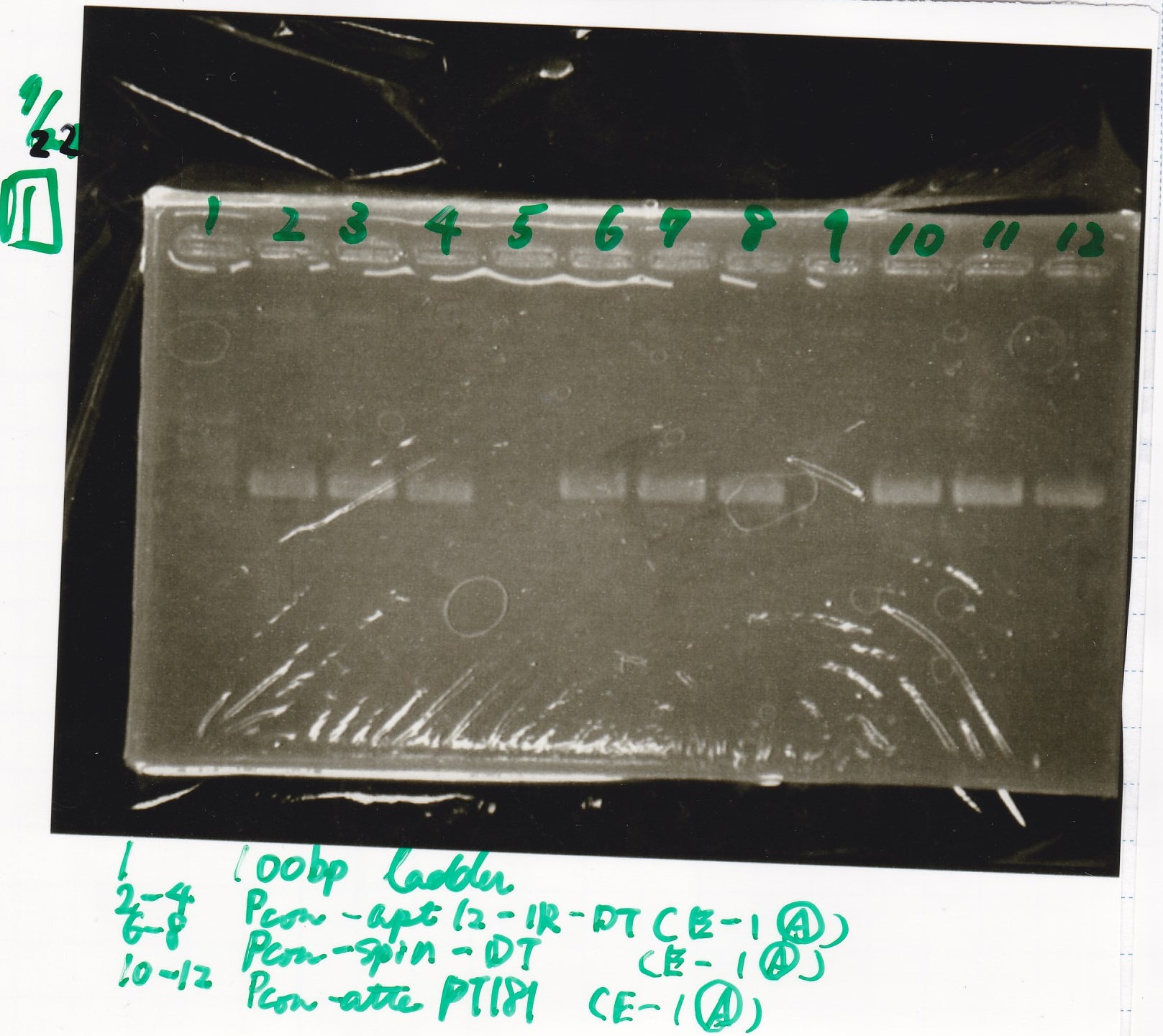
===PCR=== | ===PCR=== | ||
<div class="experiment"> | <div class="experiment"> | ||
| - | <span class="author"> | + | <span class="author">No name</span> |
{| class="wikitable" | {| class="wikitable" | ||
!Sample||base pair | !Sample||base pair | ||
|- | |- | ||
| - | | | + | |Pcon-RBS-tetR-DT(1-2(A))||-- |
|- | |- | ||
| - | |Pcon-RBS- | + | |Pcon-RBS-tetR-DT(3-2(A))||-- |
| + | |- | ||
| + | |Pcon-RBS-GFP-DT(1-S(A))||-- | ||
| + | |- | ||
| + | |Pcon-RBS-GFP-DT(2-S(A))||-- | ||
| + | |- | ||
| + | |Pcon-tetR-DT(E-2(A))||-- | ||
|} | |} | ||
{| class="wikitable" | {| class="wikitable" | ||
!PreDenature||Denature||Annealing||Extension||cycle | !PreDenature||Denature||Annealing||Extension||cycle | ||
|- | |- | ||
| - | |94 °C||94 °C|| | + | |94 °C||94 °C||57 °C||68 °C||-- |
|- | |- | ||
| - | | | + | |2 min||10 s||30 s||1min5s||30 cycles |
|} | |} | ||
</div> | </div> | ||
| + | [[File:Igku 0922 E10.jpg]]<br> | ||
| - | + | ===Electrophoresis=== | |
| - | === | + | |
<div class="experiment"> | <div class="experiment"> | ||
| - | <span class="author"> | + | <span class="author">No name</span> |
{| class="wikitable" | {| class="wikitable" | ||
| - | ! | + | !Lane||Sample |
|- | |- | ||
| - | | | + | |1||1kbp ladder |
|- | |- | ||
| - | | | + | |2||Pcon-RBS-tetR-DT(1-2Ⓐ) |
|- | |- | ||
| - | | | + | |3||Pcon-RBS-tetR-DT(3-2Ⓐ) |
|- | |- | ||
| - | | | + | |4||Pcon-RBS-GFP-DT(1-SⒶ) |
|- | |- | ||
| - | | | + | |5||Pcon-RBS-GFP-DT(2-SⒶ) |
|- | |- | ||
| - | | | + | |6||Pcon-tetR-DT(E-2Ⓐ) |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| - | < | + | [[File:Igku 0922 E10.jpg]]<br> |
Latest revision as of 07:02, 27 September 2013
Contents |
Sep 22
Colony PCR
| Sample | base pair |
|---|---|
| 9/20 Plac-PT181 attenuator(1~4) | 664 |
| 9/20 Plac-PT181 antisense(1~6) | 369 |
| 9/20 Pcon-PT181 attenuator-aptamer-DT(1~6) | 859 |
| 9/20 Pcon-PT181 antisense-spinach-DT(1~8) | 723 |
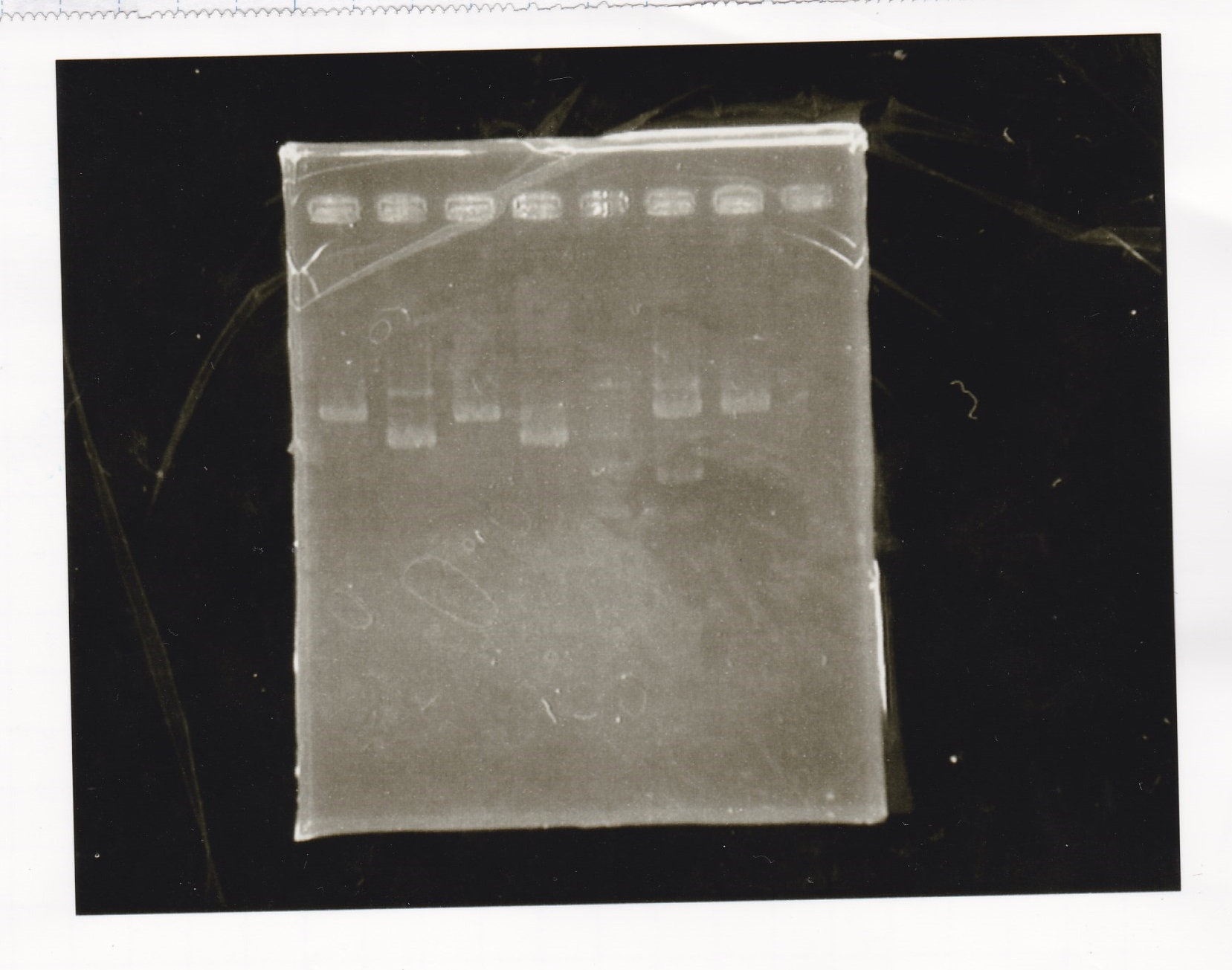
Electrophoresis
| Lane | Sample |
|---|---|
| 1 | Pcon-apt12_1R-DT(E-1A) |
| 2 | Pcon-spinach-DT(E-1A) |
| 3 | Pcon-pT181 attenuator(E-1A) |
| 4 | DT(1-3) |
| 5 | tetR aptamer 12_1R-DT(1-3) |
| 6 | 100bp ladder |
| 7 | Pcon-pT181 attenuator(1-SA) |
| 8 | spinach-DT(2-1) |
| 9 | Pcon-pT181 attenuator(E-0) |
| 10 | Pcon-pT181 attenuator(2-SA) |
| 11 | spinach-DT(3-2) |
| 12 | Ptet-PT181 anisense |
Miniprep
| DNA | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| 9/21 spinach-DT | 197.9 | 1.67 | 1.54 |
| 9/21 Plux | 125.9 | 1.71 | 1.27 |
| 9/21 Plux-RBS-GFP-DT-(1) | 168.0 | 1.62 | 1.73 |
| 9/21 Pcon-PT181 attenuator-(1) | 368.3 | 1.57 | 1.62 |
| 9/21 RBS-GFP-DT | 532.8 | 1.75 | 1.78 |
| 9/21 Pcon-RBS-GFP-DT | 845.2 | 2.02 | 1.70 |
Colony PCR
| Sample | base pair |
|---|---|
| 9/20 Plux-RBS-GFP-DT-Pcon-RBS-luxR-DT-(1~2) | 283 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 5min | 30s | 30s | 2min10s | 30cycles |
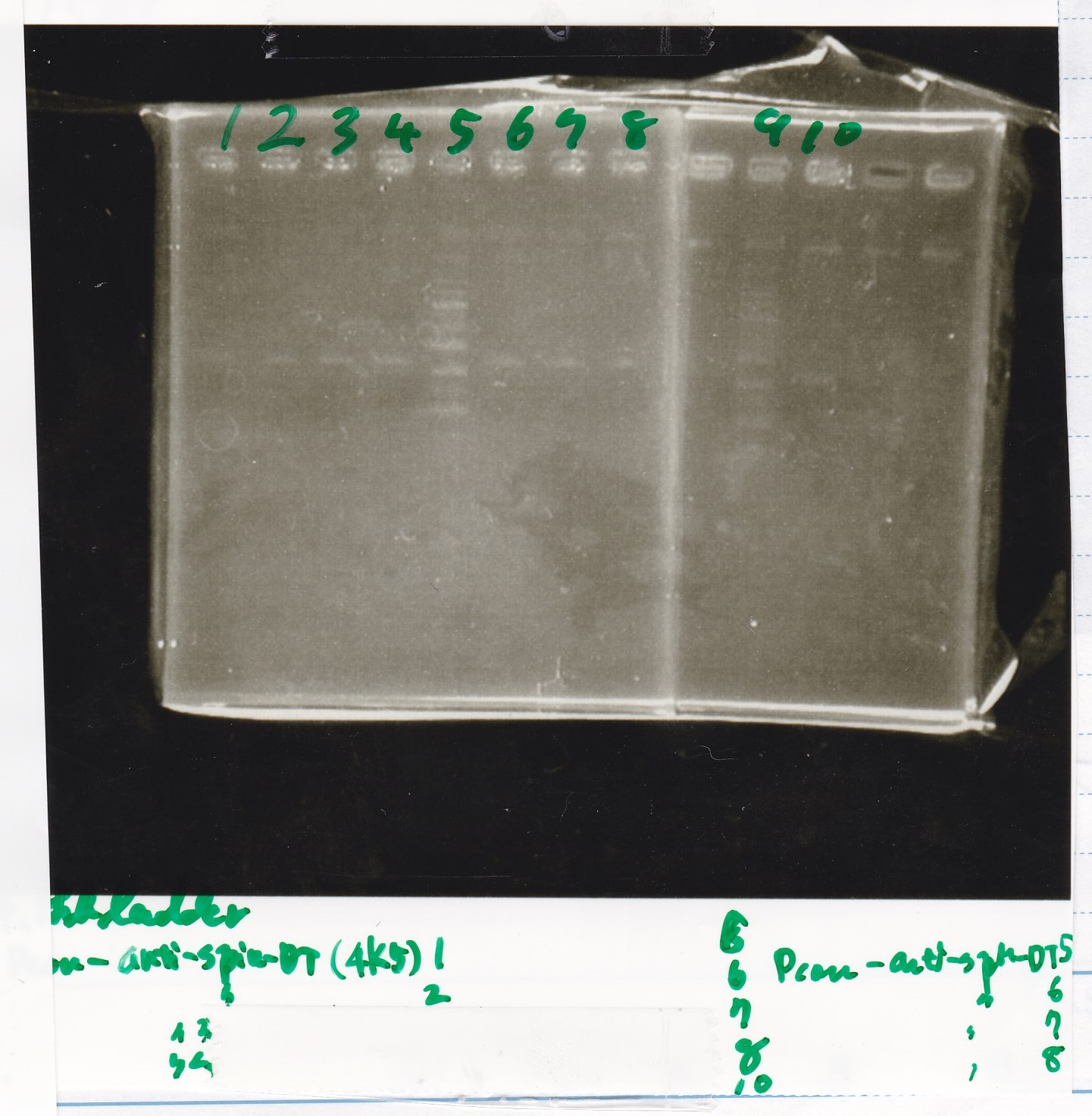
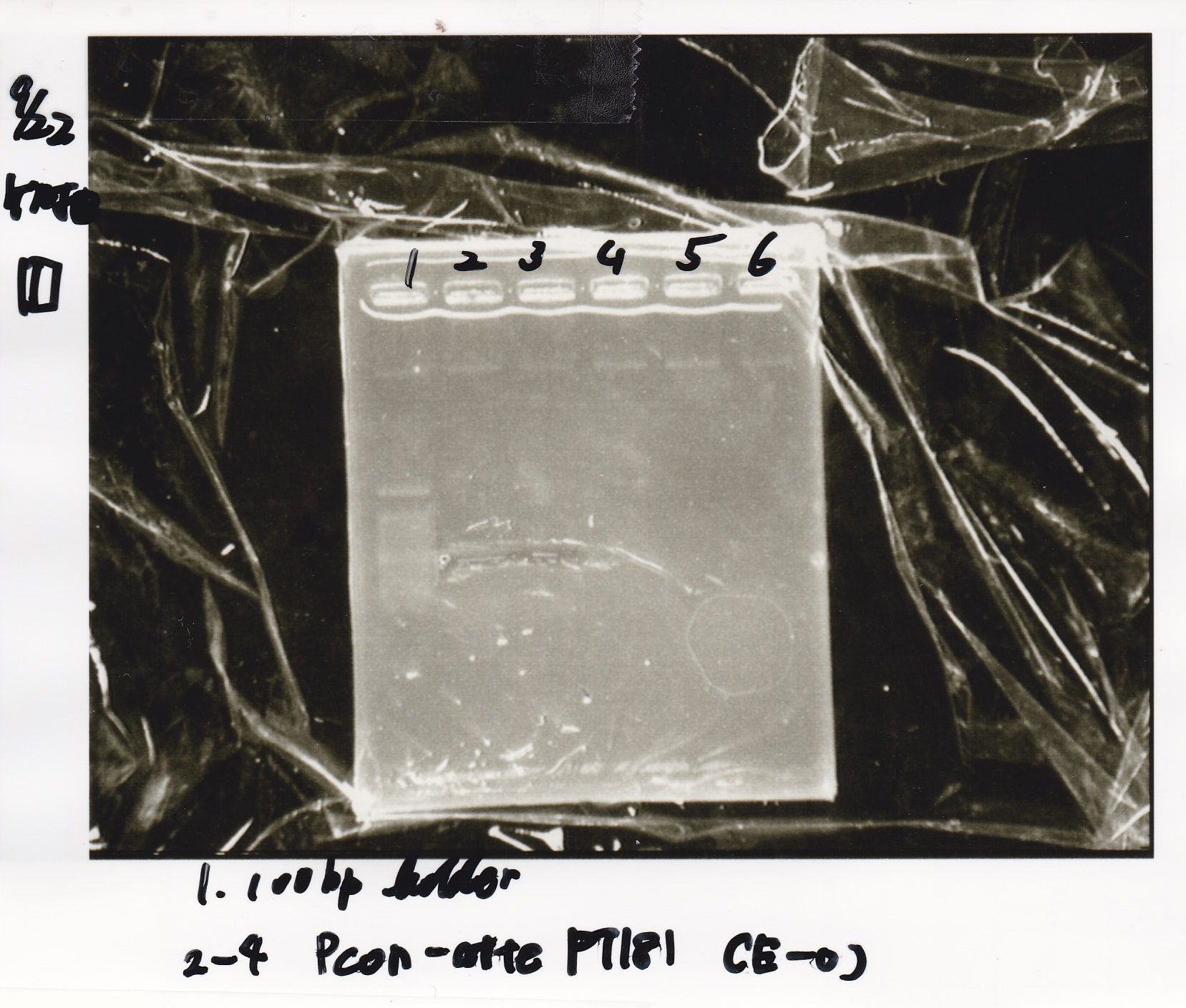
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | -- |
| 2~4 | Pcon-aptamer12-1R-DT(E-1A) | -- |
| 6~8 | Pcon-spinach-DT(E-1A) | -- |
| 10~12 | Pcon-attenuator PT181(E-1A) | -- |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | -- |
| 2~4 | DT(1-3) | -- |
| 6~8 | aptamer12-1RDT(1-3) | -- |
| 10~12 | Pcon-PT181 attenuator(1-5A) | -- |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Pcon-aptamer12-1R-DT(E-1A) | 31.2 | 1.87 | 0.30 |
| Pcon-spinach-DT(E-1A) | 25.6 | 1.88 | 0.99 |
| Pcon-attenuator PT181(E-1A) | 25.3 | 1.99 | 0.86 |
| DT(1-3) | 22.1 | 1.82 | 0.20 |
| aptamer12-1RDT(1-3) | 33.5 | 1.99 | 0.05 |
| Pcon-PT181 attenuator(1-5A) | 36.9 | 1.12 | 0.96 |
Restriction Enzyme Digestion
| 9/16 Plac | XbaI | SpeI | PstI | BufferB | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|
| 2cuts | 12.5µL | 0µL | 1µL | 1µL | 3µL | 3µL | 9.5µL | 30µL |
| NC | 0.8µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.4µL | 10µL |
| 9/22 Plux | XbaI | SpeI | PstI | BufferB | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|
| 2cuts | 16µL | 0µL | 1µL | 1µL | 3µL | 3µL | 14µL | 30µL |
| NC | 0.8µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.2µL | 10µL |
| 9/20 RBS-tetR-2R-DT | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|
| 2cuts | 8.5µL | 1µL | 0µL | 1µL | 3µL | 3µL | 13.5µL | 30µL |
| NC | 0.4µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL |
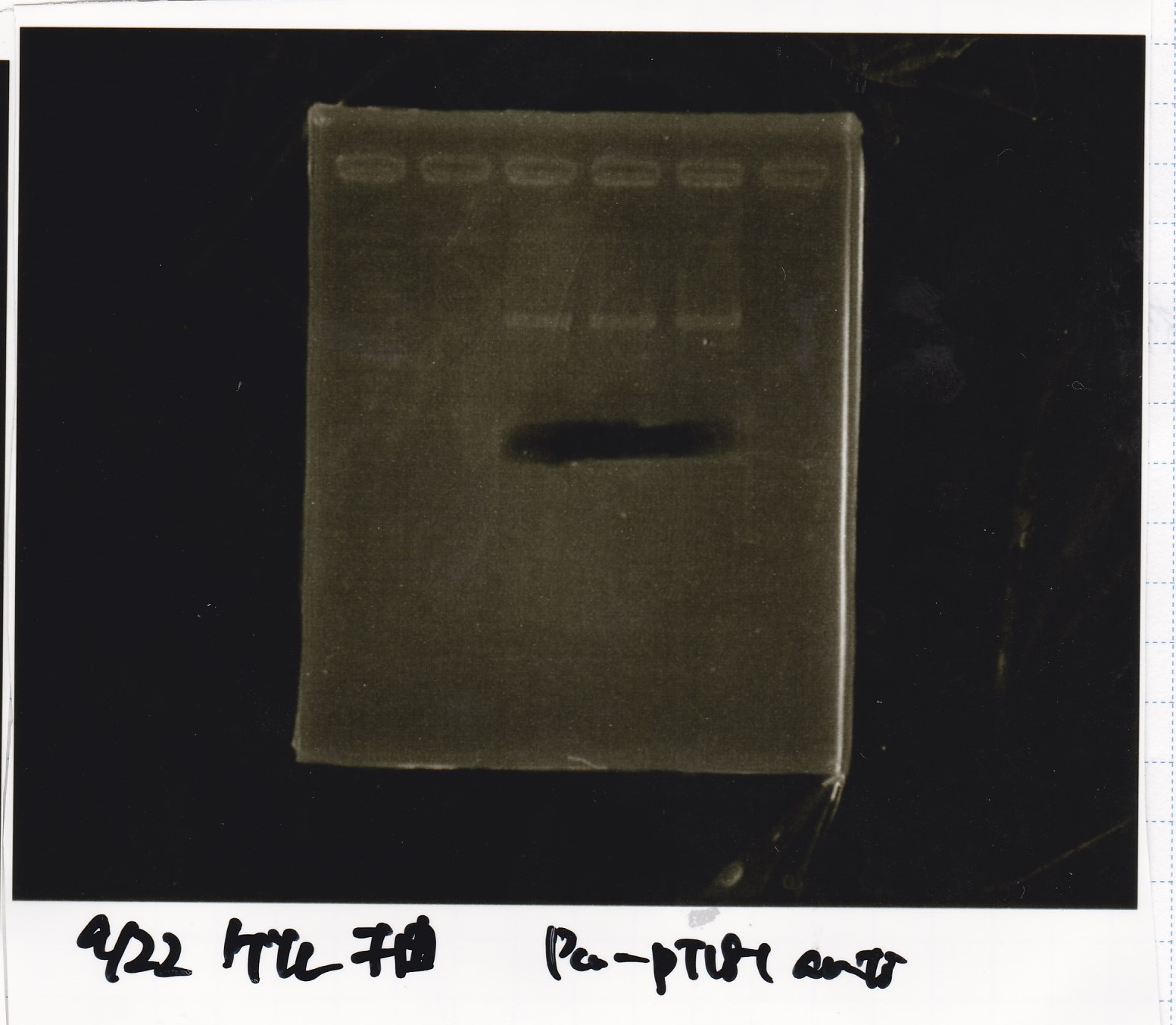
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | -- |
| 2~4 | Pcon-attenuator PT181(E-0) | -- |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | -- |
| 2~4 | Pcon-attenuator PT181(E-0) | -- |
| Lane | DNA | Enzyme |
|---|---|---|
| 2 | 100bp ladder | -- |
| 4~6 | Pcon-attenuator PT181(2-SA) | -- |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | -- |
| 3~5 | spinach-DT(3-2) | -- |
| 8~10 | Ptet-antisense PT181(1-3) | -- |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Pcon-attenuator PT181(E-0) | 2.4 | 1.80 | 0.17 |
| Pcon-attenuator PT181(2-SA) | 10.8 | 1.85 | 0.54 |
| spinach-DT(3-2) | 36.7 | 1.80 | 0.75 |
| Ptet-antisense PT181(1-3) | 16.8 | 1.74 | 0.78 |
Colony PCR
| Sample | base pair |
|---|---|
| 9/20 Pcon-PT181 attenuator-aptamer12-1R-DT-(7~14) | 859 |
Colony PCR
| Sample | base pair |
|---|---|
| PconPT181antisense-spinach-DT(9~15) | 723 |
Ligation
| state | Vector | Inserter | Ligation High ver.2 | ||
|---|---|---|---|---|---|
| experiment | 9/13 Pcon-PT181 attenuator (SpeI & PstI) | 1.7 µL | 9/18 aptamer-1R-DT (XbaI & PstI) | 5.4µL | 3.5µL |
| experiment | 9/12 Plac(SpeI & PstI) | 2.4 µL | 9/18 aptamer-1R-DT (XbaI & PstI) | 6.1µL | 3.5 µL |
| experiment | 9/13 Ptet(SpeI & PstI) | 1.7 µL | 9/10 Plux-RBS-GFP-DT | 7.0 µL | 4.4 µL |
| experiment | 9/21 RBS-GFP-DT (EcoRI & XbaI) | 0.6 µL | 9/18 aptamer 12-1R-DT (XbaI & PstI) | 5.6µL | 3.1 µL |
| experiment | 9/13 PSB1C3(XbaI & PstI) | 1.9 µL | 9/16 PT181 attenuator(XbaI & PstI) | 4.6µL | 3.3 µL |
Liquid Culture
| Sample | medium |
|---|---|
| 9/20 Plac-antisense-1 | -- |
| 9/20 Plac-attenuator-1 | -- |
| 9/20 Plac-antisense-1 | -- |
| 9/20 Plac-attenuator-1 | -- |
digestion
| 9/8 Pcon-PT181 antisense | EcoRI | SpeI | BufferM | MilliQ | total | |
|---|---|---|---|---|---|---|
| 2cuts | 8.3µL | 1µL | 1µL | 3µL | 16.7µL | 30µL |
| NC | 0.4µL | 0µL | 0µL | 1µL | 8.3µL | 10µL |
| 9/7 Pcon-RBS-tetR-DT | EcoRI | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2cut | 14µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 18.2µL | 30µL |
| NC | 0.7µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.3µL | 10µL |
| 9/22 RBS-GFP-DT | EcoRI | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2cuts | 3.8µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 18.2µL | 30µL |
| NC | 0.2µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.8µL | 10µL |
| 9/6 aptamer12-1R-DT | EcoRI | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | |
|---|---|---|---|---|---|---|---|---|
| 2cut | 8µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 16µL |
| NC | 0.4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL |
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kbp ladder | -- |
| 3~5 | Pcon-PT181 antisense (EcoRI&SpaI) | -- |
Transformation
| Name | Sample | Competent Cells | Plate |
|---|---|---|---|
| 9/20 Plac-aptamer12-1R-DT(XbaI&PstI) | 2 µL | 20 µL | Amp |
| 9/20 pT181 attenuator(1C3)(EcoRI&SpeI) | 2 µL | 20 µL | CP |
| 9/17 pT181 attenuator(1C3)(XbaI&PstI) | 2 µL | 20 µL | CP |
| 9/20 Pcon-pT181 attenuator(SpaI&PstI)+aptamer12-1R-DT(XbaI&PstI) | 2 µL | 20 µL | Amp |
| kaiABC | 2 µL | 20 µL | Amp |
| 9/22 pSB1C3-RpaB(XbaI&PstI) | 2 µL | 20 µL | Amp |
| 9/22 pSB1C3-pT181 attenuator(XbaI&PstI) | 2 µL | 20 µL | Amp |
| 9/22 Ptet(SpaI&PstI)+RBS-GFP-DT(XbaI&PstI) | 2 µL | 20 µL | CP |
| 9/22 RBS-GFP-DT+Pcon-attenuator pT181 (EcoRI&SpeI) | 2 µL | 20 µL | CP |
| 9/22 pT181 attenuator(XbaI&PstI)(1C3) | 2 µL | 20 µL | CP |
PCR
| Sample | base pair |
|---|---|
| Pcon-RBS-tetR-DT(1-2(A)) | -- |
| Pcon-RBS-tetR-DT(3-2(A)) | -- |
| Pcon-RBS-GFP-DT(1-S(A)) | -- |
| Pcon-RBS-GFP-DT(2-S(A)) | -- |
| Pcon-tetR-DT(E-2(A)) | -- |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94 °C | 94 °C | 57 °C | 68 °C | -- |
| 2 min | 10 s | 30 s | 1min5s | 30 cycles |
 "
"