Team:Shenzhen BGIC 0101/Modules
From 2013.igem.org
(Created page with "<html> <head> <!-- iGem wiki hacks --> <!-- Remove all empty <p> tags --> <script type="text/javascript"> $(document).ready(function() { ...") |
|||
| (36 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:Shenzhen BGIC 0101/Templates/Header}} | ||
<html> | <html> | ||
<head> | <head> | ||
| - | + | <hr style="color:#BDCBBD; height:3px;" /> | |
| - | + | <div id="module"> | |
| - | + | <h1>Neochr - New Chromosome Designer</h1> | |
| - | + | <p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The Neochr module contains three plugins: Decouple, Add and delete.<br/> | |
| - | + | <img class="hehe" src="https://static.igem.org/mediawiki/2013/6/6e/Decouple.png" /><br/> | |
| - | + | <img class="hehe" src="https://static.igem.org/mediawiki/2013/a/a7/Add.png" /><br/> | |
| - | + | <img class="hehe" src="https://static.igem.org/mediawiki/2013/8/8c/Delete.png" /><br/> | |
| - | + | </p> | |
| - | + | <p> | |
| + | Neochr module uses public data from <a href="http://www.yeastgenome.org">Saccharomyces Genome Database, </a> | ||
| + | <a href="http://www.genome.jp/kegg">Kyoto Encyclopedia of Genes and Genomes</a> , <a href="http://www.genome.wisc.edu">E.coli genome project</a>,<a href="http://www.mgc.ac.cn/VFs/main.htm"> Virulence Factors of Pathogenic Bacteria </a>.<br/> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/1/11/Mm3.jpg" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/5/5d/M4.jpg" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/2/2f/Mm1.jpg" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/b/bf/Mm2.jpg" /> | ||
| + | <br/><br/> | ||
| + | <hr style="color:#BDCBBD; height:3px;" /> | ||
| - | + | <h1>NucleoMod - Nucleotide Sequence Modifier</h1> | |
| - | + | <p>When we extract gene from wild type genome to create a new chromosome, We need to silence the original wild type gene. The NucleoMod module can design CRISPR site to reach this goal.And the module can optimize the codon to increase expression level of genes.</p> | |
| - | + | <p><img class="hehe" src="https://static.igem.org/mediawiki/2013/8/82/Crispr-01.png" /><br/> | |
| - | + | <img class="hehe" src="https://static.igem.org/mediawiki/2013/d/d8/Create-delete-enzyme-site.png" /><br/> | |
| - | + | <img class="hehe" src="https://static.igem.org/mediawiki/2013/7/7a/Codon-optization.png" /><br/> | |
| - | + | <img class="hehe" src="https://static.igem.org/mediawiki/2013/f/f6/Repeat_smash.png" /> | |
| - | + | </p> | |
| - | + | <hr style="color:#BDCBBD; height:3px;" /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | // | + | |
| - | + | <h1>SegmMan - Split for Assembly</h1> | |
| - | + | <p>The synthesizer or synthesis chip can up to 3kb DNA sequence with high accuracy, but chromosome is not that short.<br/> | |
| + | SegmMan can settle this problem, it splits chromosome into 30k fragments, after parsing its exited enzyme sites, continues segmentation into 10k and 2k fragments. In 10k and 2k level, its will add vector homologous region and design enzyme sites.<br/> | ||
| + | <img class="hehe" src="https://static.igem.org/mediawiki/2013/f/f9/Plugins-Work-Flow.png" /><br/><br/><br/><br/> | ||
| + | <img class="hehe" src="https://static.igem.org/mediawiki/2013/9/9d/Poster-Segman.png" /> | ||
| + | </p> | ||
| + | <br/><br/> | ||
| + | <hr style="color:#BDCBBD; height:3px;" /> | ||
| - | < | + | <h1>OLS(Oligo library synthesis) Designer</h1> |
| - | < | + | <p>For the users who synthesize DNA in chip can utilize OLS(Oligo library synthesis) Designer to design oligos and enzyme sites, primers to assemble 500-800 fragments.</p> |
| - | < | + | <p><img class="hehe" src="https://static.igem.org/mediawiki/2013/6/63/Chip_design_process.png" /></p> |
| - | + | <p>(a) Pre-designed oligonucleotides (no distinction is made between dsDNA and ssDNA in the figure) are synthesized on a DNA microchip.<br/> | |
| - | < | + | (b) cleaved to make a pool of oligo nucleotides.<br/> |
| - | + | (c) Plate-specific primer sequences (yellow or brown) are used to amplify separate plate subpools (only two areshown), which contain DNA to assemble different genes(only three are shown for each plate subpool).<br/> | |
| - | + | (d) Assembly-specific sequences (shades of blue) are used to amplify assembly subpools that contain only the DNA required to make a single gene.<br/> | |
| - | + | (e) The primer sequences are cleaved using either type IIS restriction enzymes (resulting in dsDNA) or by DpnII/USER/λ exonuclease processing (producing ssDNA).<br/> | |
| - | < | + | (f) Construction primers(shown as white and black sites flanking the full assembly) are then used in an assembly PCR reaction to build a gene from each assembly subpool.<br/> |
| - | + | </p> | |
| - | + | <br/><br/> | |
| - | + | <hr style="color:#BDCBBD; height:3px;" /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | / | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <h1>Others</h1> | ||
| + | <p>Presentation from KGML<br/> | ||
| + | This module will grab genes’ detail information in KEGG Makeup Language (KGML) file which can be downloaded in <a href="http://www.genome.jp/kegg">KEGG</a> or get through KEGG API, and it will establish a new standard for data transmission which will convert XML format into JSON format and simplify structures. Furthermore, this module will export genes’ list and its relationships. Choose one pathway and this module will visualize the pathway and rebuild it in the level of genes. | ||
| + | <p style="text-align:center;"><img class="ta" src="https://static.igem.org/mediawiki/igem.org/3/3b/Mod1.png" /></p> | ||
| + | <br/><br/> | ||
| + | <hr style="color:#BDCBBD; height:3px;" /> | ||
| + | </p> | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 15:19, 28 October 2013
Neochr - New Chromosome Designer
The Neochr module contains three plugins: Decouple, Add and delete.



Neochr module uses public data from Saccharomyces Genome Database,
Kyoto Encyclopedia of Genes and Genomes , E.coli genome project, Virulence Factors of Pathogenic Bacteria .




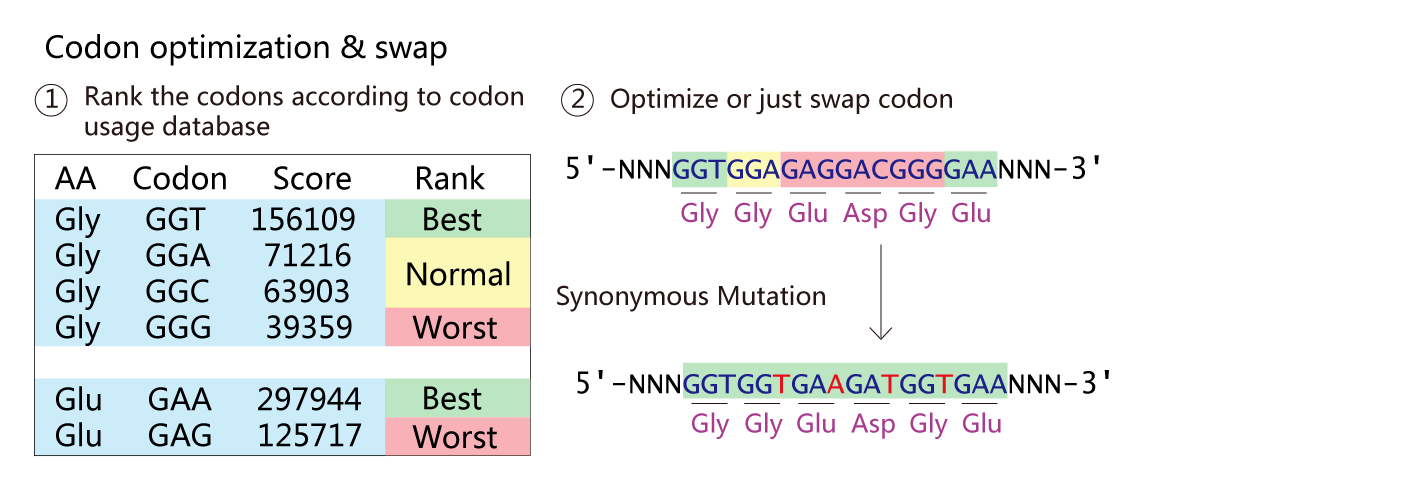
NucleoMod - Nucleotide Sequence Modifier
When we extract gene from wild type genome to create a new chromosome, We need to silence the original wild type gene. The NucleoMod module can design CRISPR site to reach this goal.And the module can optimize the codon to increase expression level of genes.



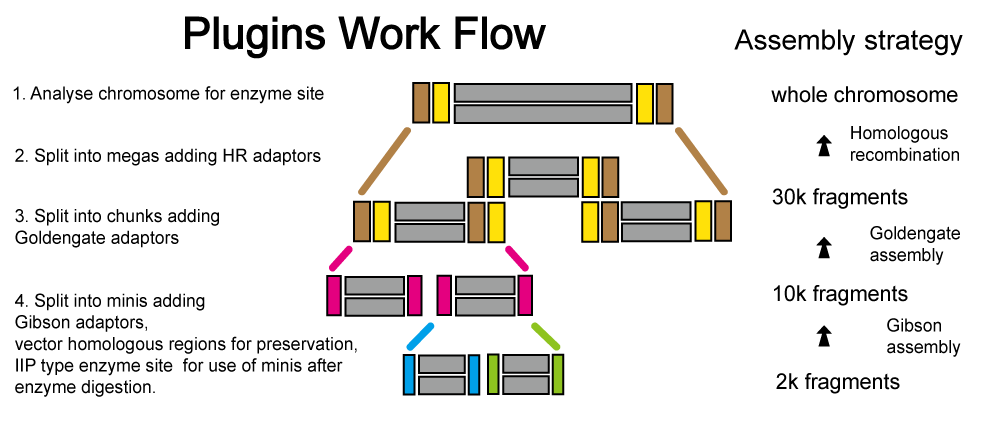
SegmMan - Split for Assembly
The synthesizer or synthesis chip can up to 3kb DNA sequence with high accuracy, but chromosome is not that short.
SegmMan can settle this problem, it splits chromosome into 30k fragments, after parsing its exited enzyme sites, continues segmentation into 10k and 2k fragments. In 10k and 2k level, its will add vector homologous region and design enzyme sites.

OLS(Oligo library synthesis) Designer
For the users who synthesize DNA in chip can utilize OLS(Oligo library synthesis) Designer to design oligos and enzyme sites, primers to assemble 500-800 fragments.

(a) Pre-designed oligonucleotides (no distinction is made between dsDNA and ssDNA in the figure) are synthesized on a DNA microchip.
(b) cleaved to make a pool of oligo nucleotides.
(c) Plate-specific primer sequences (yellow or brown) are used to amplify separate plate subpools (only two areshown), which contain DNA to assemble different genes(only three are shown for each plate subpool).
(d) Assembly-specific sequences (shades of blue) are used to amplify assembly subpools that contain only the DNA required to make a single gene.
(e) The primer sequences are cleaved using either type IIS restriction enzymes (resulting in dsDNA) or by DpnII/USER/λ exonuclease processing (producing ssDNA).
(f) Construction primers(shown as white and black sites flanking the full assembly) are then used in an assembly PCR reaction to build a gene from each assembly subpool.
Others
Presentation from KGML
This module will grab genes’ detail information in KEGG Makeup Language (KGML) file which can be downloaded in KEGG or get through KEGG API, and it will establish a new standard for data transmission which will convert XML format into JSON format and simplify structures. Furthermore, this module will export genes’ list and its relationships. Choose one pathway and this module will visualize the pathway and rebuild it in the level of genes.

 "
"



