Team:Stanford-Brown/Projects/CRISPR
From 2013.igem.org
Sophia liang (Talk | contribs) (→Acknowledgements) |
(→Future Work) |
||
| (48 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Stanford-Brown/Templates/Main}} | {{:Team:Stanford-Brown/Templates/Main}} | ||
| + | __NOTOC__ | ||
| + | == '''Accomplishments''' == | ||
| + | <ul> | ||
| + | <li>Submitted bricks for Cas9, a minimal CRISPR array, and a Cas9 variant suitable for transcriptional regulation</li> | ||
| + | <li>Demonstrated conjugation of RP4</li> | ||
| + | </ul> | ||
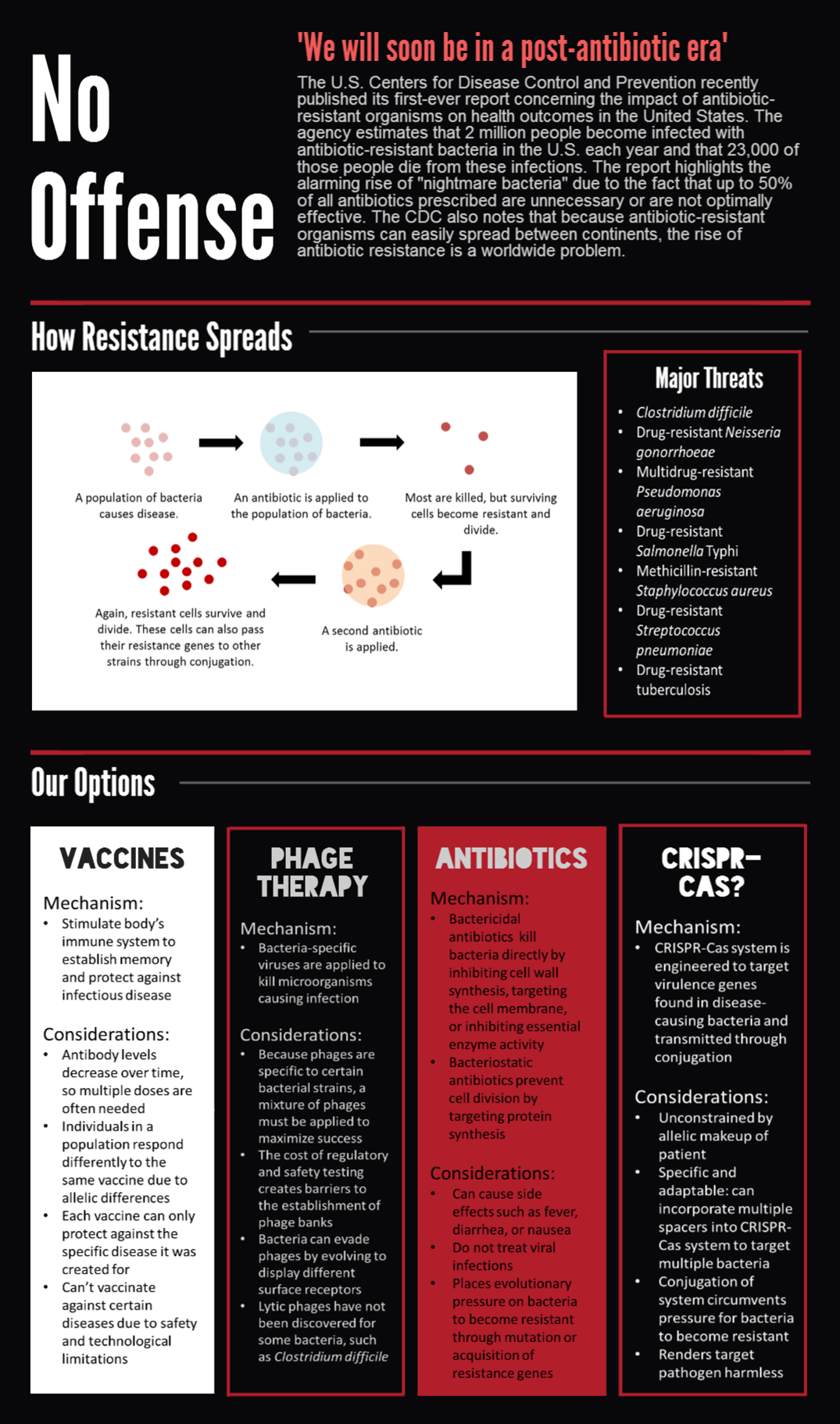
== '''Introduction''' == | == '''Introduction''' == | ||
| - | |||
| - | + | CRISPR-Cas bacterial immune systems are capable of remembering and targeting invasive DNA by storing short DNA sequences, called spacers, in genomic arrays called CRISPRs. RNA transcripts of the spacers are then incorporated into CRISPR-associated (Cas) proteins, which surveill the cell and degrade DNA complementary to the spacer. | |
| - | < | + | The most-studied CRISPR-Cas system is the Cas9 protein from <i>Streptococcus pyogenes</i>, which has drawn much attention for its utility in genome editing. We sought to BioBrick the Cas9 protein and variants for a different purpose — programmable transcriptional regulation. By using a "deactivated" Cas9 mutant dCas9 that retains DNA-binding but not degrading activity, we sought to brick a transcriptional repressor that can be directed to arbitrary regions of the genome (Bikard et al., 2013). We also sought to brick the dCas9-ω transcriptional activator, which contains the omega RNA polymerase subunit fused to the carboxy terminus of dCas9 (Bikard et al., 2013). |
| - | + | To extend on previous work using Cas9 as a transcriptional regulator, we sought to conjugate it between cells to regulate specific target genes in an entire population. We envision the system as means to combat antibiotic resistance by degrading resistance plasmids or even rendering the pathogen harmless. | |
| - | + | ||
| - | + | ||
== '''Protocols''' == | == '''Protocols''' == | ||
| - | A complete list of our protocols can be found | + | A complete list of our protocols can be found [https://docs.google.com/a/brown.edu/document/d/10hpfdvthGIWRPrrQCnWNIy3HfinFoK0hlV7VMb5nGHI '''here'''.] |
== '''Lab Notebook''' == | == '''Lab Notebook''' == | ||
| - | '''[https://docs.google.com/document/d/1BN6_bs12q0t2Isztzf2nWQl0Yfo8MOG1mkFD927RNNQ/edit?usp=sharing CRISPR-Cas | + | '''[https://docs.google.com/document/d/1BN6_bs12q0t2Isztzf2nWQl0Yfo8MOG1mkFD927RNNQ/edit?usp=sharing We documented our experimental work in an online notebook.]''' |
| + | |||
| + | Briefly, selected CRISPR-Cas components were amplified using Q5 polymerase (NEB) from clones provided by Dr. David Bikard. Primers contained 20bp of end homology for circular polymerase extension cloning into pSB1C3 (Quan ''et al.'', 2009). Site-directed mutagenesis was performed to remove an internal EcoRI site via a modified Quikchange method (Liu ''et. al'', 2008). All parts were Sanger sequenced before submission. | ||
== '''Data''' == | == '''Data''' == | ||
| - | We submitted bricks for Cas9, a minimal CRISPR array, and transcriptional activator variant of Cas9, dCas9-ω | + | We submitted bricks for Cas9, a minimal CRISPR array, and transcriptional activator variant of Cas9, dCas9-ω. Our characterization of the bricks is pending while we construct fluorescent testing constructs to demonstrate the cleaveage activity of Cas9 and regulatory activity of dCas9-ω when introduced via conjugation. |
| - | + | We also demonstrated successful conjugation of the RP4 plasmid. Electrocompetent cells were transformed with RP4, a plasmid coding for proteins needed for conjugation, and grown in liquid culture containing ampicillin. The ampicillin-resistant RP4 cells were mated with cells containing chloramphenicol-resistant RFP, and the resulting strain was plated on a double antibiotic amp+chlor plate. The two parent strains were also streaked to confirm that each did not possess the needed resistance genes prior to conjugation. | |
| - | We also demonstrated successful conjugation of the RP4 plasmid. Electrocompetent cells were transformed with RP4, a plasmid coding for proteins needed for conjugation, and grown in liquid culture containing | + | |
<br> | <br> | ||
| - | <center>[[File:Team Stanford Brown 2013 RP4.png| | + | <center> |
| + | {| class="wikitable" style="text-align: center" | ||
| + | | [[File:Team Stanford Brown 2013 RP4.png|490px]] | ||
| + | |- | ||
| + | | Figure 1. Mating experiment to prove cells harboring RP4 plasmid can conjugate. <br> | ||
| + | |} | ||
| + | </center> | ||
<br> | <br> | ||
After conjugation was confirmed, we performed a midiprep of the RP4 cells to obtain the RP4 plasmid. Later, this RP4 plasmid will be transformed into our donor cells to facilitate conjugation of the CRISPR-Cas system. | After conjugation was confirmed, we performed a midiprep of the RP4 cells to obtain the RP4 plasmid. Later, this RP4 plasmid will be transformed into our donor cells to facilitate conjugation of the CRISPR-Cas system. | ||
| + | |||
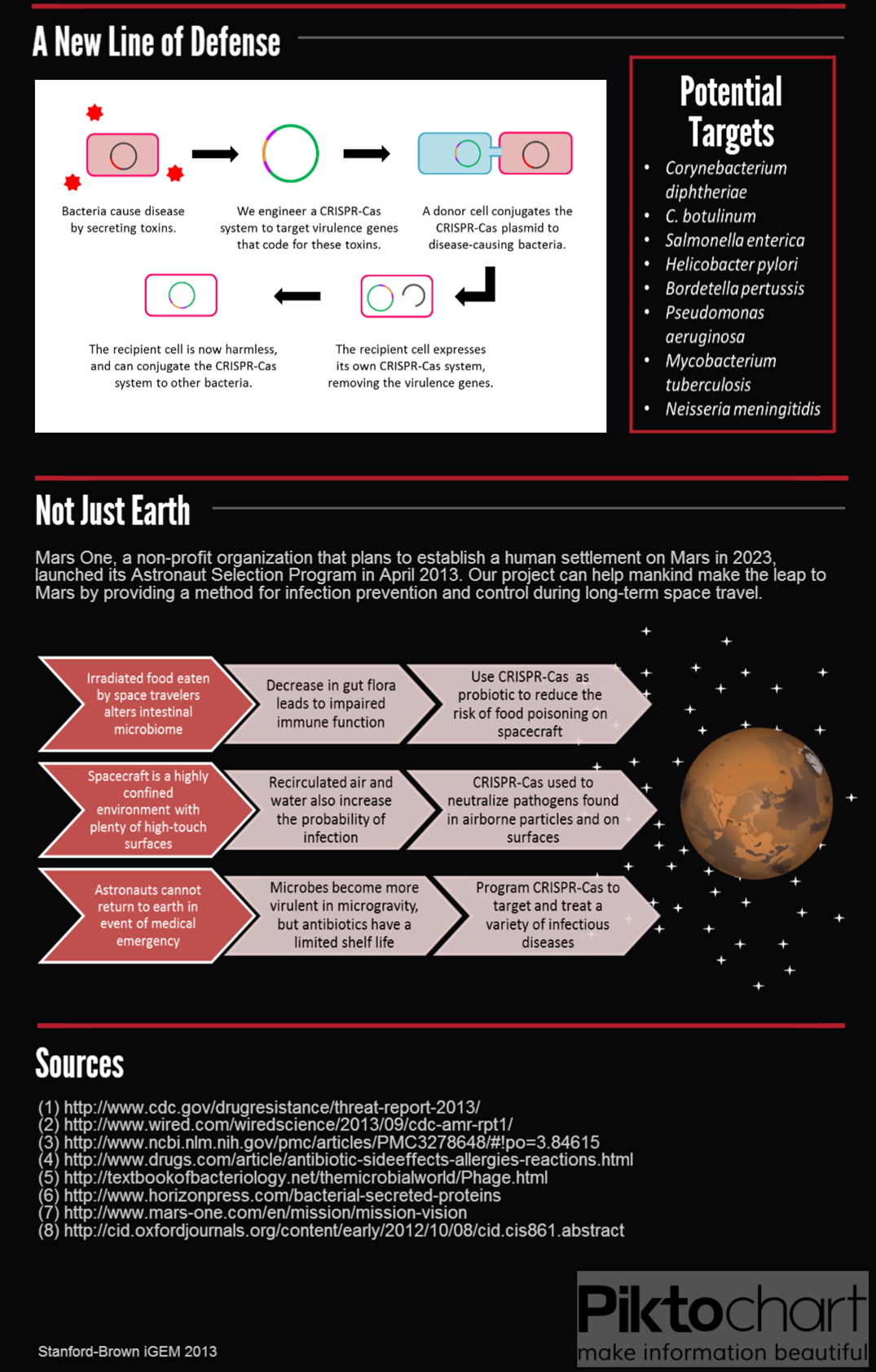
| + | == '''Applications''' == | ||
<br> | <br> | ||
| - | == | + | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">[[File:Medical&Space (4).png|753x1276px]][[File:Team Stanford Brown 2013 Medical&Space(bottomhalf).png|753x1179px]]</div> |
<br> | <br> | ||
| + | |||
<div class="center" style="width: auto; margin-left: auto; margin-right: auto;">[[File:O157 (4).png]]</div> | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">[[File:O157 (4).png]]</div> | ||
<br> | <br> | ||
| + | |||
| + | == '''Future Work''' == | ||
| + | In the future we hope to continue characterizing our system via fluorometry and fluorescent microscopy. We intend to show that receiving our Cas9 brick via conjugation results in cleavage of a target fluorescence plasmid. We also intend to show that conjugation of dCas9-ω results in transcriptional repression when directed to a promoter or coding region and transcriptional activation when directed upstream of a promoter. | ||
== '''BioBricks''' == | == '''BioBricks''' == | ||
| - | '''[http://parts.igem.org/Part: | + | '''[http://parts.igem.org/Part:BBa_K1218000 BBa_K1218000 (Minimal CRISPR Array)]''' This part codes for a minimal CRISPR array of a type II CRISPR-Cas system. The CRISPR array includes two CRISPR repeats separated by a spacer with two BsaI sites. Digestion with BsaI allows for insertion of a new spacer, thus changing the sequence targeted by Cas9. |
| - | '''[http://parts.igem.org/Part: | + | '''[http://parts.igem.org/Part:BBa_K1218011 BBa_K1218011 (pCas9)]''' This part codes for the tracrRNA, Cas9 protein, and minimal CRISPR array of a type II CRISPR-Cas system. The CRISPR array includes two CRISPR repeats separated by a spacer with two BsaI sites. Digestion with BsaI allows for insertion of a new spacer, thus changing the sequence targeted by Cas9. |
| - | '''[http://parts.igem.org/Part:BBa_K1218014 | + | '''[http://parts.igem.org/Part:BBa_K1218014 BBa_K1218014 (dCas9-ω Activator)]''' This part codes for the tracrRNA and a mutated Cas9 protein. It is designed to be used with BBa_K1218000. It carries two mutations abolishing DNA cleavage activity while preserving DNA binding, and it is fused to the RNA polymerase omega subunit on its carboxylic acid terminus. Binding of dCas9-ω upstream of a promoter activates transcription of the targeted gene. |
| - | == '''Acknowledgements'''== | + | == '''Acknowledgements''' == |
We would like to thank: | We would like to thank: | ||
<ul> | <ul> | ||
| - | <li>Dr. Lynn Rothschild, Dr. Gary Wessel, and Dr. Joe Shih | + | <li>Dr. Lynn Rothschild, Dr. Gary Wessel, and Dr. Joe Shih - our primary advisors</li> |
| - | <li>Dr. David | + | <li>Dr. David Bikard - for providing Cas9 plasmids and clarifying questions about promoter choice</li> |
| + | <li>Dr. Leonard A. Mermel - whose paper provided one of the primary inspirations for this project</li> | ||
| + | <li>Dr. Kosuke Fujisima - for mentoring us in the lab</li> | ||
| + | <li>Jane Berry at EMD Millipore </li> | ||
| + | <li>Coli Genomic Stock Center [CGSC]</li> | ||
</ul> | </ul> | ||
| + | |||
| + | == '''References''' == | ||
| + | |||
| + | *Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., & Marraffini, L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Research. | ||
| + | *Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., ... & Charpentier, E. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature, 471(7340), 602-607. | ||
| + | *Jiang, W., Bikard, D., Cox, D., Zhang, F., & Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature biotechnology, 31(3), 233-239. | ||
| + | *Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science, 337(6096), 816-821. | ||
| + | *Liu, H., & Naismith, J. H. (2008). An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC biotechnology, 8(1), 91. | ||
| + | *Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., ... & Church, G. M. (2013). RNA-guided human genome engineering via Cas9. Science, 339(6121), 823-826. | ||
| + | *Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., & Lim, W. A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 152(5), 1173-1183. | ||
| + | *Quan, J., & Tian, J. (2009). Circular polymerase extension cloning of complex gene libraries and pathways. PloS one, 4(7), e6441. | ||
| + | *Wang, H., Yang, H., Shivalila, C. S., Dawlaty, M. M., Cheng, A. W., Zhang, F., & Jaenisch, R. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. | ||
Latest revision as of 03:31, 29 October 2013
Accomplishments
- Submitted bricks for Cas9, a minimal CRISPR array, and a Cas9 variant suitable for transcriptional regulation
- Demonstrated conjugation of RP4
Introduction
CRISPR-Cas bacterial immune systems are capable of remembering and targeting invasive DNA by storing short DNA sequences, called spacers, in genomic arrays called CRISPRs. RNA transcripts of the spacers are then incorporated into CRISPR-associated (Cas) proteins, which surveill the cell and degrade DNA complementary to the spacer.
The most-studied CRISPR-Cas system is the Cas9 protein from Streptococcus pyogenes, which has drawn much attention for its utility in genome editing. We sought to BioBrick the Cas9 protein and variants for a different purpose — programmable transcriptional regulation. By using a "deactivated" Cas9 mutant dCas9 that retains DNA-binding but not degrading activity, we sought to brick a transcriptional repressor that can be directed to arbitrary regions of the genome (Bikard et al., 2013). We also sought to brick the dCas9-ω transcriptional activator, which contains the omega RNA polymerase subunit fused to the carboxy terminus of dCas9 (Bikard et al., 2013).
To extend on previous work using Cas9 as a transcriptional regulator, we sought to conjugate it between cells to regulate specific target genes in an entire population. We envision the system as means to combat antibiotic resistance by degrading resistance plasmids or even rendering the pathogen harmless.
Protocols
A complete list of our protocols can be found here.
Lab Notebook
We documented our experimental work in an online notebook.
Briefly, selected CRISPR-Cas components were amplified using Q5 polymerase (NEB) from clones provided by Dr. David Bikard. Primers contained 20bp of end homology for circular polymerase extension cloning into pSB1C3 (Quan et al., 2009). Site-directed mutagenesis was performed to remove an internal EcoRI site via a modified Quikchange method (Liu et. al, 2008). All parts were Sanger sequenced before submission.
Data
We submitted bricks for Cas9, a minimal CRISPR array, and transcriptional activator variant of Cas9, dCas9-ω. Our characterization of the bricks is pending while we construct fluorescent testing constructs to demonstrate the cleaveage activity of Cas9 and regulatory activity of dCas9-ω when introduced via conjugation.
We also demonstrated successful conjugation of the RP4 plasmid. Electrocompetent cells were transformed with RP4, a plasmid coding for proteins needed for conjugation, and grown in liquid culture containing ampicillin. The ampicillin-resistant RP4 cells were mated with cells containing chloramphenicol-resistant RFP, and the resulting strain was plated on a double antibiotic amp+chlor plate. The two parent strains were also streaked to confirm that each did not possess the needed resistance genes prior to conjugation.
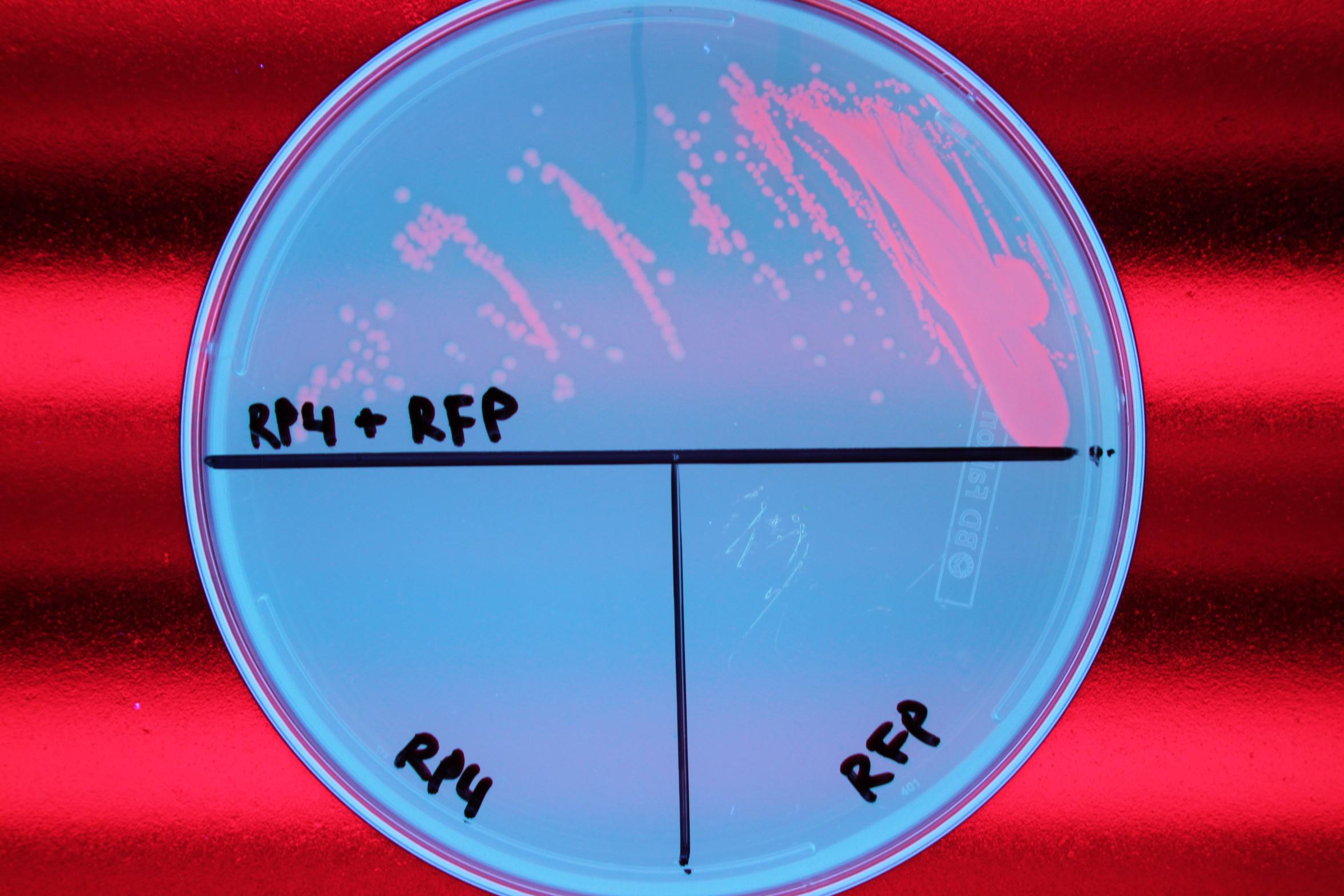
|
| Figure 1. Mating experiment to prove cells harboring RP4 plasmid can conjugate. |
After conjugation was confirmed, we performed a midiprep of the RP4 cells to obtain the RP4 plasmid. Later, this RP4 plasmid will be transformed into our donor cells to facilitate conjugation of the CRISPR-Cas system.
Applications
Future Work
In the future we hope to continue characterizing our system via fluorometry and fluorescent microscopy. We intend to show that receiving our Cas9 brick via conjugation results in cleavage of a target fluorescence plasmid. We also intend to show that conjugation of dCas9-ω results in transcriptional repression when directed to a promoter or coding region and transcriptional activation when directed upstream of a promoter.
BioBricks
[http://parts.igem.org/Part:BBa_K1218000 BBa_K1218000 (Minimal CRISPR Array)] This part codes for a minimal CRISPR array of a type II CRISPR-Cas system. The CRISPR array includes two CRISPR repeats separated by a spacer with two BsaI sites. Digestion with BsaI allows for insertion of a new spacer, thus changing the sequence targeted by Cas9.
[http://parts.igem.org/Part:BBa_K1218011 BBa_K1218011 (pCas9)] This part codes for the tracrRNA, Cas9 protein, and minimal CRISPR array of a type II CRISPR-Cas system. The CRISPR array includes two CRISPR repeats separated by a spacer with two BsaI sites. Digestion with BsaI allows for insertion of a new spacer, thus changing the sequence targeted by Cas9.
[http://parts.igem.org/Part:BBa_K1218014 BBa_K1218014 (dCas9-ω Activator)] This part codes for the tracrRNA and a mutated Cas9 protein. It is designed to be used with BBa_K1218000. It carries two mutations abolishing DNA cleavage activity while preserving DNA binding, and it is fused to the RNA polymerase omega subunit on its carboxylic acid terminus. Binding of dCas9-ω upstream of a promoter activates transcription of the targeted gene.
Acknowledgements
We would like to thank:
- Dr. Lynn Rothschild, Dr. Gary Wessel, and Dr. Joe Shih - our primary advisors
- Dr. David Bikard - for providing Cas9 plasmids and clarifying questions about promoter choice
- Dr. Leonard A. Mermel - whose paper provided one of the primary inspirations for this project
- Dr. Kosuke Fujisima - for mentoring us in the lab
- Jane Berry at EMD Millipore
- Coli Genomic Stock Center [CGSC]
References
- Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., & Marraffini, L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Research.
- Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., ... & Charpentier, E. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature, 471(7340), 602-607.
- Jiang, W., Bikard, D., Cox, D., Zhang, F., & Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature biotechnology, 31(3), 233-239.
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science, 337(6096), 816-821.
- Liu, H., & Naismith, J. H. (2008). An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC biotechnology, 8(1), 91.
- Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., ... & Church, G. M. (2013). RNA-guided human genome engineering via Cas9. Science, 339(6121), 823-826.
- Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., & Lim, W. A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 152(5), 1173-1183.
- Quan, J., & Tian, J. (2009). Circular polymerase extension cloning of complex gene libraries and pathways. PloS one, 4(7), e6441.
- Wang, H., Yang, H., Shivalila, C. S., Dawlaty, M. M., Cheng, A. W., Zhang, F., & Jaenisch, R. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell.
 "
"