Team:UC-Santa Cruz/Project/Overview
From 2013.igem.org
Hacking.sean (Talk | contribs) |
|||
| (18 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:UC-SantaCruz_Template}} | {{:Team:UC-SantaCruz_Template}} | ||
| + | <html xmlns:o="urn:schemas-microsoft-com:office:office" | ||
| + | xmlns:w="urn:schemas-microsoft-com:office:word" | ||
| + | xmlns:m="http://schemas.microsoft.com/office/2004/12/omml" | ||
| + | xmlns="http://www.w3.org/TR/REC-html40"> | ||
| - | + | <head> | |
| - | + | <meta name=Title content=""> | |
| - | 2 | + | <meta name=Keywords content=""> |
| - | + | <meta http-equiv=Content-Type content="text/html; charset=macintosh"> | |
| - | + | <meta name=ProgId content=Word.Document> | |
| - | 5. | + | <meta name=Generator content="Microsoft Word 2008"> |
| - | 6. | + | <meta name=Originator content="Microsoft Word 2008"> |
| + | <link rel=File-List href="Project%20Overview_9_27_13_files/filelist.xml"> | ||
| + | <!--[if gte mso 9]><xml> | ||
| + | <o:DocumentProperties> | ||
| + | <o:Author>Hugh Olsen</o:Author> | ||
| + | <o:Template>Normal.dotm</o:Template> | ||
| + | <o:LastAuthor>Hugh Olsen</o:LastAuthor> | ||
| + | <o:Revision>2</o:Revision> | ||
| + | <o:TotalTime>212</o:TotalTime> | ||
| + | <o:Created>2013-09-27T21:02:00Z</o:Created> | ||
| + | <o:LastSaved>2013-09-27T21:02:00Z</o:LastSaved> | ||
| + | <o:Pages>1</o:Pages> | ||
| + | <o:Words>659</o:Words> | ||
| + | <o:Characters>3758</o:Characters> | ||
| + | <o:Lines>31</o:Lines> | ||
| + | <o:Paragraphs>7</o:Paragraphs> | ||
| + | <o:CharactersWithSpaces>4615</o:CharactersWithSpaces> | ||
| + | <o:Version>12.0</o:Version> | ||
| + | </o:DocumentProperties> | ||
| + | <o:OfficeDocumentSettings> | ||
| + | <o:AllowPNG/> | ||
| + | </o:OfficeDocumentSettings> | ||
| + | </xml><![endif]--><!--[if gte mso 9]><xml> | ||
| + | <w:WordDocument> | ||
| + | <w:SpellingState>Clean</w:SpellingState> | ||
| + | <w:GrammarState>Clean</w:GrammarState> | ||
| + | <w:TrackMoves>false</w:TrackMoves> | ||
| + | <w:TrackFormatting/> | ||
| + | <w:PunctuationKerning/> | ||
| + | <w:DrawingGridHorizontalSpacing>18 pt</w:DrawingGridHorizontalSpacing> | ||
| + | <w:DrawingGridVerticalSpacing>18 pt</w:DrawingGridVerticalSpacing> | ||
| + | <w:DisplayHorizontalDrawingGridEvery>0</w:DisplayHorizontalDrawingGridEvery> | ||
| + | <w:DisplayVerticalDrawingGridEvery>0</w:DisplayVerticalDrawingGridEvery> | ||
| + | <w:ValidateAgainstSchemas/> | ||
| + | <w:SaveIfXMLInvalid>false</w:SaveIfXMLInvalid> | ||
| + | <w:IgnoreMixedContent>false</w:IgnoreMixedContent> | ||
| + | <w:AlwaysShowPlaceholderText>false</w:AlwaysShowPlaceholderText> | ||
| + | <w:Compatibility> | ||
| + | <w:BreakWrappedTables/> | ||
| + | <w:DontGrowAutofit/> | ||
| + | <w:DontAutofitConstrainedTables/> | ||
| + | <w:DontVertAlignInTxbx/> | ||
| + | </w:Compatibility> | ||
| + | </w:WordDocument> | ||
| + | </xml><![endif]--><!--[if gte mso 9]><xml> | ||
| + | <w:LatentStyles DefLockedState="false" LatentStyleCount="276"> | ||
| + | </w:LatentStyles> | ||
| + | </xml><![endif]--> | ||
| + | <style> | ||
| + | <!-- | ||
| + | /* Font Definitions */ | ||
| + | @font-face | ||
| + | {font-family:"Courier New"; | ||
| + | panose-1:2 7 3 9 2 2 5 2 4 4; | ||
| + | mso-font-charset:0; | ||
| + | mso-generic-font-family:auto; | ||
| + | mso-font-pitch:variable; | ||
| + | mso-font-signature:3 0 0 0 1 0;} | ||
| + | @font-face | ||
| + | {font-family:Wingdings; | ||
| + | panose-1:5 2 1 2 1 8 4 8 7 8; | ||
| + | mso-font-charset:2; | ||
| + | mso-generic-font-family:auto; | ||
| + | mso-font-pitch:variable; | ||
| + | mso-font-signature:0 0 65536 0 -2147483648 0;} | ||
| + | @font-face | ||
| + | {font-family:Cambria; | ||
| + | panose-1:2 4 5 3 5 4 6 3 2 4; | ||
| + | mso-font-charset:0; | ||
| + | mso-generic-font-family:auto; | ||
| + | mso-font-pitch:variable; | ||
| + | mso-font-signature:3 0 0 0 1 0;} | ||
| + | /* Style Definitions */ | ||
| + | p.MsoNormal, li.MsoNormal, div.MsoNormal | ||
| + | {mso-style-parent:""; | ||
| + | margin:0in; | ||
| + | margin-bottom:.0001pt; | ||
| + | mso-pagination:widow-orphan; | ||
| + | font-size:12.0pt; | ||
| + | font-family:"Times New Roman"; | ||
| + | mso-ascii-font-family:Cambria; | ||
| + | mso-ascii-theme-font:minor-latin; | ||
| + | mso-fareast-font-family:Cambria; | ||
| + | mso-fareast-theme-font:minor-latin; | ||
| + | mso-hansi-font-family:Cambria; | ||
| + | mso-hansi-theme-font:minor-latin; | ||
| + | mso-bidi-font-family:"Times New Roman"; | ||
| + | mso-bidi-theme-font:minor-bidi;} | ||
| + | p.MsoListParagraph, li.MsoListParagraph, div.MsoListParagraph | ||
| + | {margin-top:0in; | ||
| + | margin-right:0in; | ||
| + | margin-bottom:0in; | ||
| + | margin-left:.5in; | ||
| + | margin-bottom:.0001pt; | ||
| + | mso-add-space:auto; | ||
| + | mso-pagination:widow-orphan; | ||
| + | font-size:12.0pt; | ||
| + | font-family:"Times New Roman"; | ||
| + | mso-ascii-font-family:Cambria; | ||
| + | mso-ascii-theme-font:minor-latin; | ||
| + | mso-fareast-font-family:Cambria; | ||
| + | mso-fareast-theme-font:minor-latin; | ||
| + | mso-hansi-font-family:Cambria; | ||
| + | mso-hansi-theme-font:minor-latin; | ||
| + | mso-bidi-font-family:"Times New Roman"; | ||
| + | mso-bidi-theme-font:minor-bidi;} | ||
| + | p.MsoListParagraphCxSpFirst, li.MsoListParagraphCxSpFirst, div.MsoListParagraphCxSpFirst | ||
| + | {mso-style-type:export-only; | ||
| + | margin-top:0in; | ||
| + | margin-right:0in; | ||
| + | margin-bottom:0in; | ||
| + | margin-left:.5in; | ||
| + | margin-bottom:.0001pt; | ||
| + | mso-add-space:auto; | ||
| + | mso-pagination:widow-orphan; | ||
| + | font-size:12.0pt; | ||
| + | font-family:"Times New Roman"; | ||
| + | mso-ascii-font-family:Cambria; | ||
| + | mso-ascii-theme-font:minor-latin; | ||
| + | mso-fareast-font-family:Cambria; | ||
| + | mso-fareast-theme-font:minor-latin; | ||
| + | mso-hansi-font-family:Cambria; | ||
| + | mso-hansi-theme-font:minor-latin; | ||
| + | mso-bidi-font-family:"Times New Roman"; | ||
| + | mso-bidi-theme-font:minor-bidi;} | ||
| + | p.MsoListParagraphCxSpMiddle, li.MsoListParagraphCxSpMiddle, div.MsoListParagraphCxSpMiddle | ||
| + | {mso-style-type:export-only; | ||
| + | margin-top:0in; | ||
| + | margin-right:0in; | ||
| + | margin-bottom:0in; | ||
| + | margin-left:.5in; | ||
| + | margin-bottom:.0001pt; | ||
| + | mso-add-space:auto; | ||
| + | mso-pagination:widow-orphan; | ||
| + | font-size:12.0pt; | ||
| + | font-family:"Times New Roman"; | ||
| + | mso-ascii-font-family:Cambria; | ||
| + | mso-ascii-theme-font:minor-latin; | ||
| + | mso-fareast-font-family:Cambria; | ||
| + | mso-fareast-theme-font:minor-latin; | ||
| + | mso-hansi-font-family:Cambria; | ||
| + | mso-hansi-theme-font:minor-latin; | ||
| + | mso-bidi-font-family:"Times New Roman"; | ||
| + | mso-bidi-theme-font:minor-bidi;} | ||
| + | p.MsoListParagraphCxSpLast, li.MsoListParagraphCxSpLast, div.MsoListParagraphCxSpLast | ||
| + | {mso-style-type:export-only; | ||
| + | margin-top:0in; | ||
| + | margin-right:0in; | ||
| + | margin-bottom:0in; | ||
| + | margin-left:.5in; | ||
| + | margin-bottom:.0001pt; | ||
| + | mso-add-space:auto; | ||
| + | mso-pagination:widow-orphan; | ||
| + | font-size:12.0pt; | ||
| + | font-family:"Times New Roman"; | ||
| + | mso-ascii-font-family:Cambria; | ||
| + | mso-ascii-theme-font:minor-latin; | ||
| + | mso-fareast-font-family:Cambria; | ||
| + | mso-fareast-theme-font:minor-latin; | ||
| + | mso-hansi-font-family:Cambria; | ||
| + | mso-hansi-theme-font:minor-latin; | ||
| + | mso-bidi-font-family:"Times New Roman"; | ||
| + | mso-bidi-theme-font:minor-bidi;} | ||
| + | span.SpellE | ||
| + | {mso-style-name:""; | ||
| + | mso-spl-e:yes;} | ||
| + | span.GramE | ||
| + | {mso-style-name:""; | ||
| + | mso-gram-e:yes;} | ||
| + | @page Section1 | ||
| + | {size:8.5in 11.0in; | ||
| + | margin:1.0in 1.25in 1.0in 1.25in; | ||
| + | mso-header-margin:.5in; | ||
| + | mso-footer-margin:.5in; | ||
| + | mso-paper-source:0;} | ||
| + | div.Section1 | ||
| + | {page:Section1;} | ||
| + | /* List Definitions */ | ||
| + | @list l0 | ||
| + | {mso-list-id:1; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:1 1 2 -1 -1 -1 -1 -1 -1 -1;} | ||
| + | @list l0:level1 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l0:level2 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l0:level3 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l0:level4 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l0:level5 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l0:level6 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l0:level7 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l0:level8 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l0:level9 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l1 | ||
| + | {mso-list-id:2; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:2 101 -1 -1 -1 -1 -1 -1 -1 -1;} | ||
| + | @list l1:level1 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:\25AA; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l1:level2 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l1:level3 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l1:level4 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l1:level5 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l1:level6 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l1:level7 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l1:level8 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l1:level9 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l2 | ||
| + | {mso-list-id:3; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:3 201 -1 -1 -1 -1 -1 -1 -1 -1;} | ||
| + | @list l2:level1 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:\25AA; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l2:level2 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l2:level3 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l2:level4 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l2:level5 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l2:level6 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l2:level7 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l2:level8 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l2:level9 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l3 | ||
| + | {mso-list-id:4; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:4 301 -1 -1 -1 -1 -1 -1 -1 -1;} | ||
| + | @list l3:level1 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:\25AA; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l3:level2 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l3:level3 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l3:level4 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l3:level5 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l3:level6 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l3:level7 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l3:level8 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l3:level9 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l4 | ||
| + | {mso-list-id:5; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:5 401 -1 -1 -1 -1 -1 -1 -1 -1;} | ||
| + | @list l4:level1 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:\25AA; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l4:level2 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l4:level3 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l4:level4 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l4:level5 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l4:level6 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l4:level7 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l4:level8 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l4:level9 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l5 | ||
| + | {mso-list-id:6; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:6 501 -1 -1 -1 -1 -1 -1 -1 -1;} | ||
| + | @list l5:level1 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:\25AA; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l5:level2 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l5:level3 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l5:level4 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l5:level5 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l5:level6 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l5:level7 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l5:level8 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l5:level9 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l6 | ||
| + | {mso-list-id:7; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:7 601 -1 -1 -1 -1 -1 -1 -1 -1;} | ||
| + | @list l6:level1 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:\25AA; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l6:level2 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l6:level3 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l6:level4 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l6:level5 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l6:level6 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l6:level7 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l6:level8 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l6:level9 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l7 | ||
| + | {mso-list-id:8; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:8 701 -1 -1 -1 -1 -1 -1 -1 -1;} | ||
| + | @list l7:level1 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:\25AA; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l7:level2 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l7:level3 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l7:level4 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l7:level5 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l7:level6 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l7:level7 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l7:level8 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l7:level9 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l8 | ||
| + | {mso-list-id:9; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:9 801 -1 -1 -1 -1 -1 -1 -1 -1;} | ||
| + | @list l8:level1 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:\25AA; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l8:level2 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l8:level3 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l8:level4 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l8:level5 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l8:level6 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l8:level7 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l8:level8 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l8:level9 | ||
| + | {mso-level-start-at:0; | ||
| + | mso-level-text:""; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | margin-left:0in; | ||
| + | text-indent:0in;} | ||
| + | @list l9 | ||
| + | {mso-list-id:682820741; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:1012194386 67698689 67698691 67698693 67698689 67698691 67698693 67698689 67698691 67698693;} | ||
| + | @list l9:level1 | ||
| + | {mso-level-number-format:bullet; | ||
| + | mso-level-text:\F0B7; | ||
| + | mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in; | ||
| + | font-family:Symbol;} | ||
| + | @list l10 | ||
| + | {mso-list-id:931162519; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:1082415826 67698703 67698691 67698693 67698689 67698691 67698693 67698689 67698691 67698693;} | ||
| + | @list l10:level1 | ||
| + | {mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | @list l11 | ||
| + | {mso-list-id:1386876092; | ||
| + | mso-list-type:hybrid; | ||
| + | mso-list-template-ids:1082415826 67698703 67698691 67698693 67698689 67698691 67698693 67698689 67698691 67698693;} | ||
| + | @list l11:level1 | ||
| + | {mso-level-tab-stop:none; | ||
| + | mso-level-number-position:left; | ||
| + | text-indent:-.25in;} | ||
| + | ol | ||
| + | {margin-bottom:0in;} | ||
| + | ul | ||
| + | {margin-bottom:0in;} | ||
| + | --> | ||
| + | </style> | ||
| + | <!--[if gte mso 10]> | ||
| + | <style> | ||
| + | /* Style Definitions */ | ||
| + | table.MsoNormalTable | ||
| + | {mso-style-name:"Table Normal"; | ||
| + | mso-tstyle-rowband-size:0; | ||
| + | mso-tstyle-colband-size:0; | ||
| + | mso-style-noshow:yes; | ||
| + | mso-style-parent:""; | ||
| + | mso-padding-alt:0in 5.4pt 0in 5.4pt; | ||
| + | mso-para-margin:0in; | ||
| + | mso-para-margin-bottom:.0001pt; | ||
| + | mso-pagination:widow-orphan; | ||
| + | font-size:12.0pt; | ||
| + | font-family:"Times New Roman"; | ||
| + | mso-ascii-font-family:Cambria; | ||
| + | mso-ascii-theme-font:minor-latin; | ||
| + | mso-hansi-font-family:Cambria; | ||
| + | mso-hansi-theme-font:minor-latin;} | ||
| + | </style> | ||
| + | <![endif]--> | ||
| + | </head> | ||
| - | + | <body bgcolor=white lang=EN-US style='tab-interval:.5in'> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <div class=Section1> | ||
| - | + | <p class=MsoNormal style='margin-bottom:6.0pt;mso-pagination:none;mso-layout-grid-align: | |
| - | + | none;text-autospace:none'><span style='font-size:19.0pt;font-family:Helvetica; | |
| - | + | mso-bidi-font-family:Helvetica'>ABSTRACT<o:p></o:p></span></p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | <p class=MsoNormal style='margin-bottom:6.0pt;mso-pagination:none;mso-layout-grid-align: | ||
| + | none;text-autospace:none'><span style='font-size:13.0pt;font-family:Helvetica; | ||
| + | mso-bidi-font-family:Helvetica'>
Fresh water shortages affect half of the | ||
| + | world’s population (currently 7 billion persons). In the next 25 years the | ||
| + | numbers of people impacted by <u>severe</u> water shortages is expected to | ||
| + | increase four fold. Shortage of potable water has been estimated to account for | ||
| + | 80-90% of disease and 30% of mortality for humanity. <span class=GramE>Roughly | ||
| + | half of all accessible fresh waters (rivers, lakes and underground aquifers) | ||
| + | are estimated to be in use by the current world population</span>. Given only 2.5 | ||
| + | percent of the total volume of water is fresh water and this percentage is | ||
| + | shrinking with global warming, fresh water is becoming a critical limiting | ||
| + | resource for human populations. The goal of this project is to develop an | ||
| + | efficient, green, scalable, low cost solution for desalinization of <span | ||
| + | class=GramE>sea water</span> and brackish water. Our solution is creation of a <span | ||
| + | class=SpellE>biofilm</span> (<span class=SpellE>biomachine</span><span | ||
| + | class=GramE>) which</span> uses sunlight to pump sodium chloride across the <span | ||
| + | class=SpellE>biofilm</span> for the desalinization of salt and brackish waters, essentially replacing the pump, membrane, and energy source of a reverse osmosis machine. | ||
| + | The project also provides a scaffold for possible <span class=SpellE>biofilm</span> | ||
| + | cleanup of heavy metals and other toxic ionic pollutants.<o:p></o:p></span></p> | ||
| - | + | <p class=MsoNormal style='margin-bottom:11.0pt;mso-pagination:none;mso-layout-grid-align: | |
| + | none;text-autospace:none'><span style='font-size:19.0pt;font-family:Helvetica; | ||
| + | mso-bidi-font-family:Helvetica'>Project Goals<o:p></o:p></span></p> | ||
| - | + | <p class=MsoListParagraphCxSpFirst style='margin-bottom:11.0pt;mso-add-space: | |
| - | + | auto;text-indent:-.25in;mso-pagination:none;mso-list:l10 level1 lfo11; | |
| - | + | mso-layout-grid-align:none;text-autospace:none'><![if !supportLists]><span | |
| - | + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-fareast-font-family: | |
| - | + | Helvetica;mso-bidi-font-family:Helvetica'><span style='mso-list:Ignore'>1.<span | |
| - | + | style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span | |
| - | + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | |
| + | Helvetica'>Identify a prototype <span class=SpellE>polarizable</span> | ||
| + | prokaryotic <span class=SpellE>biofilm</span> species<o:p></o:p></span></p> | ||
| - | + | <p class=MsoListParagraphCxSpMiddle style='margin-bottom:11.0pt;mso-add-space: | |
| - | + | auto;text-indent:-.25in;mso-pagination:none;mso-list:l10 level1 lfo11; | |
| - | + | mso-layout-grid-align:none;text-autospace:none'><![if !supportLists]><span | |
| - | 2. | + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-fareast-font-family: |
| - | + | Helvetica;mso-bidi-font-family:Helvetica'><span style='mso-list:Ignore'>2.<span | |
| - | + | style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span | |
| - | + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | |
| - | + | Helvetica'>Identify and Clone <span class=GramE>protein tagging</span> peptides | |
| + | for intracellular targeting of pumps and channels to specific cellular poles.<o:p></o:p></span></p> | ||
| - | + | <p class=MsoListParagraphCxSpMiddle style='margin-bottom:11.0pt;mso-add-space: | |
| - | + | auto;text-indent:-.25in;mso-pagination:none;mso-list:l10 level1 lfo11; | |
| - | + | mso-layout-grid-align:none;text-autospace:none'><![if !supportLists]><span | |
| - | + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-fareast-font-family: | |
| - | + | Helvetica;mso-bidi-font-family:Helvetica'><span style='mso-list:Ignore'>3.<span | |
| - | + | style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span | |
| - | 7. | + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: |
| - | + | Helvetica'>Identify and Clone a series of sunlight driven ion pumps (<span | |
| - | + | class=SpellE>halorhodopsins</span>)<o:p></o:p></span></p> | |
| + | <p class=MsoListParagraphCxSpMiddle style='margin-bottom:11.0pt;mso-add-space: | ||
| + | auto;text-indent:-.25in;mso-pagination:none;mso-list:l10 level1 lfo11; | ||
| + | mso-layout-grid-align:none;text-autospace:none'><![if !supportLists]><span | ||
| + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-fareast-font-family: | ||
| + | Helvetica;mso-bidi-font-family:Helvetica'><span style='mso-list:Ignore'>4.<span | ||
| + | style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span | ||
| + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'>Identify and Clone a series of ion channels<o:p></o:p></span></p> | ||
| + | <p class=MsoListParagraphCxSpLast style='margin-bottom:11.0pt;mso-add-space: | ||
| + | auto;text-indent:-.25in;mso-pagination:none;mso-list:l10 level1 lfo11; | ||
| + | mso-layout-grid-align:none;text-autospace:none'><![if !supportLists]><span | ||
| + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-fareast-font-family: | ||
| + | Helvetica;mso-bidi-font-family:Helvetica'><span style='mso-list:Ignore'>5.<span | ||
| + | style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span | ||
| + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'>Assemble appropriate tag-pump/channel constructs for assessing <span | ||
| + | class=SpellE>NaCl</span> movement across the <span class=SpellE>biofilm</span>.<o:p></o:p></span></p> | ||
| - | + | <p class=MsoNormal style='margin-bottom:11.0pt;mso-pagination:none;mso-layout-grid-align: | |
| - | + | none;text-autospace:none'><span style='font-size:19.0pt;font-family:Helvetica; | |
| - | + | mso-bidi-font-family:Helvetica'>Experimental Approach<o:p></o:p></span></p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p class=MsoListParagraphCxSpFirst style='margin-bottom:6.0pt;mso-add-space: | |
| - | + | auto;text-indent:-.25in;mso-pagination:none;mso-list:l11 level1 lfo12; | |
| - | + | mso-layout-grid-align:none;text-autospace:none'><![if !supportLists]><b | |
| - | + | style='mso-bidi-font-weight:normal'><span style='font-size:13.0pt;font-family: | |
| - | + | Helvetica;mso-fareast-font-family:Helvetica;mso-bidi-font-family:Helvetica'><span | |
| - | + | style='mso-list:Ignore'>1.<span style='font:7.0pt "Times New Roman"'> | |
| - | + | </span></span></span></b><![endif]><b style='mso-bidi-font-weight:normal'><span | |
| - | + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | |
| - | + | Helvetica'>Identify a prototype <span class=SpellE>polarizable</span> | |
| - | + | prokaryotic <span class=SpellE>biofilm</span> species. </span></b><span | |
| - | + | style='font-size:13.0pt;font-family:Helvetica;mso-bidi-font-family:Helvetica'>Our | |
| - | + | project aims to create a microbial desalination system using the bacteria <span | |
| + | class=SpellE><i style='mso-bidi-font-style:normal'>Caulobacter</i></span><i | ||
| + | style='mso-bidi-font-style:normal'> <span class=SpellE>crescentus</span></i>. <span | ||
| + | class=SpellE><i style='mso-bidi-font-style:normal'>Caulobacter</i></span><i | ||
| + | style='mso-bidi-font-style:normal'> <span class=SpellE>crescentus</span></i> | ||
| + | was chosen in our initial experiments for five reasons- 1) it is non-pathogenic | ||
| + | and widely distributed in the fresh water aquatic environment; 2) it is | ||
| + | polarized, making it possible to orient ion pumps and channels on specific sides | ||
| + | (poles) of the bacteria; 3) the stalk allows <span class=SpellE><i | ||
| + | style='mso-bidi-font-style:normal'>Caulobacter</i></span> to attach itself to | ||
| + | solid surfaces where it forms a monolayer of cells (<span class=SpellE>biofilm</span>); | ||
| + | 4) a set of plasmids is available for transformation of <span class=SpellE><i | ||
| + | style='mso-bidi-font-style:normal'>Caulobacter</i></span>; and 5) experimental fluorescent | ||
| + | tagged protein models are available for investigation of protein tagging | ||
| + | mechanisms of pole formation (stalk and flagellum) in <span class=SpellE><i | ||
| + | style='mso-bidi-font-style:normal'>Caulobacter</i></span>. Please see the | ||
| + | following key references-<b style='mso-bidi-font-weight:normal'><o:p></o:p></b></span></p> | ||
| + | <p class=MsoListParagraphCxSpMiddle style='margin-bottom:6.0pt;mso-add-space: | ||
| + | auto;mso-pagination:none;mso-layout-grid-align:none;text-autospace:none'><span | ||
| + | class=GramE><span style='mso-bidi-font-size:19.0pt;font-family:Helvetica; | ||
| + | mso-bidi-font-family:Helvetica'>Transcriptional Profiling of <span | ||
| + | class=SpellE>Caulobacter</span> <span class=SpellE>crescentus</span> during | ||
| + | Growth on Complex and Minimal Media.</span></span><span style='mso-bidi-font-size: | ||
| + | 19.0pt;font-family:Helvetica;mso-bidi-font-family:Helvetica'> <span | ||
| + | class=SpellE>Hottes</span> et al. Journal of Bacteriology 186(5)<span | ||
| + | class=GramE>:1448</span>-1461(2004).</span><span style='font-size:13.0pt; | ||
| + | font-family:Helvetica;mso-bidi-font-family:Helvetica'><o:p></o:p></span></p> | ||
| + | <p class=MsoListParagraphCxSpMiddle style='margin-bottom:11.0pt;mso-add-space: | ||
| + | auto;text-indent:-.25in;mso-pagination:none;mso-list:l11 level1 lfo12; | ||
| + | mso-layout-grid-align:none;text-autospace:none'><![if !supportLists]><b | ||
| + | style='mso-bidi-font-weight:normal'><span style='mso-bidi-font-size:19.0pt; | ||
| + | font-family:Helvetica;mso-fareast-font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'><span style='mso-list:Ignore'>2.<span style='font:7.0pt "Times New Roman"'> | ||
| + | </span></span></span></b><![endif]><b style='mso-bidi-font-weight:normal'><span | ||
| + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'>Identify and Clone <span class=GramE>protein tagging</span> peptides | ||
| + | for intracellular targeting of pumps and channels to specific cellular poles. </span></b><span | ||
| + | style='font-size:13.0pt;font-family:Helvetica;mso-bidi-font-family:Helvetica'>A | ||
| + | series of protein tags have been identified in pole formation in <span | ||
| + | class=SpellE><i style='mso-bidi-font-style:normal'>Caulobacter</i></span>. | ||
| + | These include <span class=SpellE>DivJ</span>, <span class=SpellE>TipF</span>, <span | ||
| + | class=SpellE>StpX</span>, and <span class=SpellE>PflI</span>. Cloning of these | ||
| + | tags are currently underway. Please see the following key references-</span><b | ||
| + | style='mso-bidi-font-weight:normal'><span style='mso-bidi-font-size:19.0pt; | ||
| + | font-family:Helvetica;mso-bidi-font-family:Helvetica'><o:p></o:p></span></b></p> | ||
| + | <p class=MsoListParagraphCxSpMiddle style='margin-bottom:11.0pt;mso-add-space: | ||
| + | auto;mso-pagination:none;mso-layout-grid-align:none;text-autospace:none'><span | ||
| + | class=SpellE><span class=GramE><span style='mso-bidi-font-size:19.0pt; | ||
| + | font-family:Helvetica;mso-bidi-font-family:Helvetica'>PflI</span></span></span><span | ||
| + | class=GramE><span style='mso-bidi-font-size:19.0pt;font-family:Helvetica; | ||
| + | mso-bidi-font-family:Helvetica'>, a Protein Involved in <span class=SpellE>Flagellar</span> | ||
| + | Positioning in <span class=SpellE>Caulobacter</span>.</span></span><span | ||
| + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'> <span class=SpellE>Obuchowski</span> and Jacobs-Wagner. <i | ||
| + | style='mso-bidi-font-style:normal'>Journal of Bacteriology</i> 190(5)<span | ||
| + | class=GramE>:1718</span>-1729 (2008).<o:p></o:p></span></p> | ||
| + | <p class=MsoListParagraphCxSpMiddle style='margin-bottom:11.0pt;mso-add-space: | ||
| + | auto;mso-pagination:none;mso-layout-grid-align:none;text-autospace:none'><span | ||
| + | class=GramE><span style='mso-bidi-font-size:19.0pt;font-family:Helvetica; | ||
| + | mso-bidi-font-family:Helvetica'>Protein Sequences and Cellular Factors Required | ||
| + | for Polar Localization of <span class=SpellE>Histidine</span> <span | ||
| + | class=SpellE>Kinase</span> in <span class=SpellE><i style='mso-bidi-font-style: | ||
| + | normal'>Caulobacter</i></span><i style='mso-bidi-font-style:normal'> <span | ||
| + | class=SpellE>crescentus</span></i>.</span></span><span style='mso-bidi-font-size: | ||
| + | 19.0pt;font-family:Helvetica;mso-bidi-font-family:Helvetica'><span | ||
| + | style="mso-spacerun: yes"> </span><span class=SpellE>Sciochetti</span> et | ||
| + | al., Journal of Bacteriology 184(21)<span class=GramE>:6037</span>-6049 (2002).<o:p></o:p></span></p> | ||
| - | + | <p class=MsoListParagraphCxSpMiddle style='margin-bottom:11.0pt;mso-add-space: | |
| - | + | auto;mso-pagination:none;mso-layout-grid-align:none;text-autospace:none'><span | |
| - | + | class=GramE><span style='mso-bidi-font-size:19.0pt;font-family:Helvetica; | |
| - | + | mso-bidi-font-family:Helvetica'>Protein localization and <span class=SpellE>dymanics</span> | |
| - | + | within a bacterial organelle.</span></span><span style='mso-bidi-font-size: | |
| - | + | 19.0pt;font-family:Helvetica;mso-bidi-font-family:Helvetica'> Hughes et al. | |
| - | + | PNAS<span class=GramE>:107</span>(12):559-5604(2010).<o:p></o:p></span></p> | |
| + | <p class=MsoListParagraphCxSpMiddle style='margin-bottom:11.0pt;mso-add-space: | ||
| + | auto;text-indent:-.25in;mso-pagination:none;mso-list:l11 level1 lfo12; | ||
| + | mso-layout-grid-align:none;text-autospace:none'><![if !supportLists]><b | ||
| + | style='mso-bidi-font-weight:normal'><span style='mso-bidi-font-size:19.0pt; | ||
| + | font-family:Helvetica;mso-fareast-font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'><span style='mso-list:Ignore'>3.<span style='font:7.0pt "Times New Roman"'> | ||
| + | </span></span></span></b><![endif]><b style='mso-bidi-font-weight:normal'><span | ||
| + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'>Identify and Clone a series of sunlight driven ion pumps (<span | ||
| + | class=SpellE>halorhodopsins</span>). </span></b><span style='mso-bidi-font-size: | ||
| + | 19.0pt;font-family:Helvetica;mso-bidi-font-family:Helvetica'>Candidate channels | ||
| + | include the <span class=SpellE>halorhodopsins</span> and <span class=SpellE>bacterorhodopsins</span>. | ||
| + | We have cloned one new Na pumping <span class=SpellE>halorhodopsin</span>, KR2, | ||
| + | a protein from a marine <span class=SpellE>flavobacterium</span>, <span | ||
| + | class=SpellE>Krokinobacter</span> <span class=SpellE>eikastus</span>. </span><span | ||
| + | style='font-size:13.0pt;font-family:Helvetica;mso-bidi-font-family:Helvetica'>Please | ||
| + | see the following key reference- <span class=GramE>A</span> light-driven sodium | ||
| + | ion pump in marine bacteria. Inoue et al. Nature Communications DOI: 10.1038/ncomms2689 | ||
| + | April 9, 2013. </span><span style='mso-bidi-font-size:19.0pt;font-family:Helvetica; | ||
| + | mso-bidi-font-family:Helvetica'>We are investigating function of BBaK559000 a <span | ||
| + | class=SpellE>BioBrick</span> containing a <span class=SpellE>halorhodopsin</span>. | ||
| + | We are also investigating other sources of <span class=SpellE>halorhodopsins</span>.<b | ||
| + | style='mso-bidi-font-weight:normal'><o:p></o:p></b></span></p> | ||
| - | + | <p class=MsoListParagraphCxSpMiddle style='margin-bottom:11.0pt;mso-add-space: | |
| - | + | auto;text-indent:-.25in;mso-pagination:none;mso-list:l11 level1 lfo12; | |
| + | mso-layout-grid-align:none;text-autospace:none'><![if !supportLists]><b | ||
| + | style='mso-bidi-font-weight:normal'><span style='mso-bidi-font-size:19.0pt; | ||
| + | font-family:Helvetica;mso-fareast-font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'><span style='mso-list:Ignore'>4.<span style='font:7.0pt "Times New Roman"'> | ||
| + | </span></span></span></b><![endif]><b style='mso-bidi-font-weight:normal'><span | ||
| + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'>Identify and Clone a series of ion channels. </span></b><span | ||
| + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'>We have not yet cloned any ion channels. These proteins are our next | ||
| + | goal for gene cloning.<b style='mso-bidi-font-weight:normal'><o:p></o:p></b></span></p> | ||
| - | + | <p class=MsoListParagraphCxSpLast style='margin-bottom:11.0pt;mso-add-space: | |
| - | + | auto;text-indent:-.25in;mso-pagination:none;mso-list:l11 level1 lfo12; | |
| - | + | mso-layout-grid-align:none;text-autospace:none'><![if !supportLists]><b | |
| - | + | style='mso-bidi-font-weight:normal'><span style='mso-bidi-font-size:19.0pt; | |
| - | + | font-family:Helvetica;mso-fareast-font-family:Helvetica;mso-bidi-font-family: | |
| - | + | Helvetica'><span style='mso-list:Ignore'>5.<span style='font:7.0pt "Times New Roman"'> | |
| + | </span></span></span></b><![endif]><b style='mso-bidi-font-weight:normal'><span | ||
| + | style='mso-bidi-font-size:19.0pt;font-family:Helvetica;mso-bidi-font-family: | ||
| + | Helvetica'>Assemble appropriate tag-pump/channel constructs for assessing <span | ||
| + | class=SpellE>NaCl</span> movement across the <span class=SpellE>biofilm</span>. | ||
| + | </span></b><span style='mso-bidi-font-size:19.0pt;font-family:Helvetica; | ||
| + | mso-bidi-font-family:Helvetica'>Work is in progress for creation of | ||
| + | transforming plasmids coding for fusion <span class=GramE>proteins which</span> | ||
| + | include the targeting peptide, the pump or channel and a fluorescent tag. These | ||
| + | plasmids will be used to transform <span class=SpellE><i style='mso-bidi-font-style: | ||
| + | normal'>Caulobacter</i></span><i style='mso-bidi-font-style:normal'>. </i><span | ||
| + | class=GramE>Function will be assessed by determining sub-cellular localization | ||
| + | of the fluorescent tag and assessment for sodium and chloride flux across the <span | ||
| + | class=SpellE>biofilm</span></span>.<b style='mso-bidi-font-weight:normal'><o:p></o:p></b></span></p> | ||
| - | + | <p class=MsoNormal style='mso-pagination:none;mso-layout-grid-align:none; | |
| - | + | text-autospace:none'><b><span style='font-size:17.0pt;font-family:Helvetica; | |
| - | + | mso-bidi-font-family:Helvetica'><o:p> </o:p></span></b></p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p class=MsoNormal style='margin-bottom:5.0pt;mso-pagination:none;mso-layout-grid-align: | |
| - | + | none;text-autospace:none'><span style='font-size:17.0pt;font-family:Helvetica; | |
| - | + | mso-bidi-font-family:Helvetica;mso-bidi-font-weight:bold'>For Experimental Results, | |
| + | see: <span style="mso-spacerun: yes"> </span><o:p></o:p></span></p> | ||
| - | + | <p class=MsoNormal style='margin-bottom:5.0pt;mso-pagination:none;mso-layout-grid-align: | |
| - | + | none;text-autospace:none'><span style='font-size:17.0pt;font-family:Helvetica; | |
| - | : | + | mso-bidi-font-family:Helvetica;mso-bidi-font-weight:bold'>Project <o:p></o:p></span></p> |
| + | <p class=MsoNormal style='margin-bottom:5.0pt;mso-pagination:none;mso-layout-grid-align: | ||
| + | none;text-autospace:none'><span style='font-size:17.0pt;font-family:Helvetica; | ||
| + | mso-bidi-font-family:Helvetica;mso-bidi-font-weight:bold'>Cell polarization<o:p></o:p></span></p> | ||
| - | + | <p class=MsoNormal style='margin-bottom:5.0pt;mso-pagination:none;mso-layout-grid-align: | |
| - | + | none;text-autospace:none'><span style='font-size:17.0pt;font-family:Helvetica; | |
| - | + | mso-bidi-font-family:Helvetica;mso-bidi-font-weight:bold'>Ion Pumps<o:p></o:p></span></p> | |
| - | + | ||
| - | + | <p class=MsoNormal style='margin-bottom:5.0pt;mso-pagination:none;mso-layout-grid-align: | |
| - | + | none;text-autospace:none'><span class=SpellE><span style='font-size:17.0pt; | |
| - | + | font-family:Helvetica;mso-bidi-font-family:Helvetica;mso-bidi-font-weight:bold'>BioFilm</span></span><span | |
| - | : | + | style='font-size:17.0pt;font-family:Helvetica;mso-bidi-font-family:Helvetica; |
| + | mso-bidi-font-weight:bold'><o:p></o:p></span></p> | ||
| - | + | <p class=MsoNormal style='margin-bottom:5.0pt;mso-pagination:none;mso-layout-grid-align: | |
| - | + | none;text-autospace:none'><span style='font-size:13.0pt;font-family:Helvetica; | |
| - | + | mso-bidi-font-family:Helvetica'><o:p> </o:p></span></p> | |
| - | : | + | |
| - | + | <p class=MsoNormal style='margin-bottom:5.0pt;mso-pagination:none;mso-layout-grid-align: | |
| - | + | none;text-autospace:none'><span style='font-size:17.0pt;font-family:Helvetica; | |
| - | + | mso-bidi-font-family:Helvetica;mso-bidi-font-weight:bold'><o:p> </o:p></span></p> | |
| - | : | + | |
| + | <p class=MsoNormal><o:p> </o:p></p> | ||
| - | + | </div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | </body> | |
| - | + | ||
| - | + | </html> | |
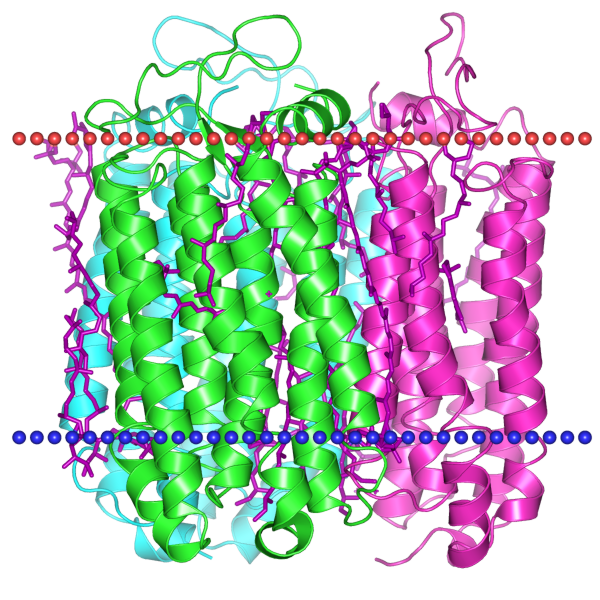
| - | + | Halorhodopsin chloride pump, viewed side-on in the cellular membrane (dotted lines) | |
| - | + | [[File:Halorhodopsin.png|400px|center]] | |
| - | + | Credit: Michael Kolbe et. al (Science, 2000) http://opm.phar.umich.edu/ | |
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
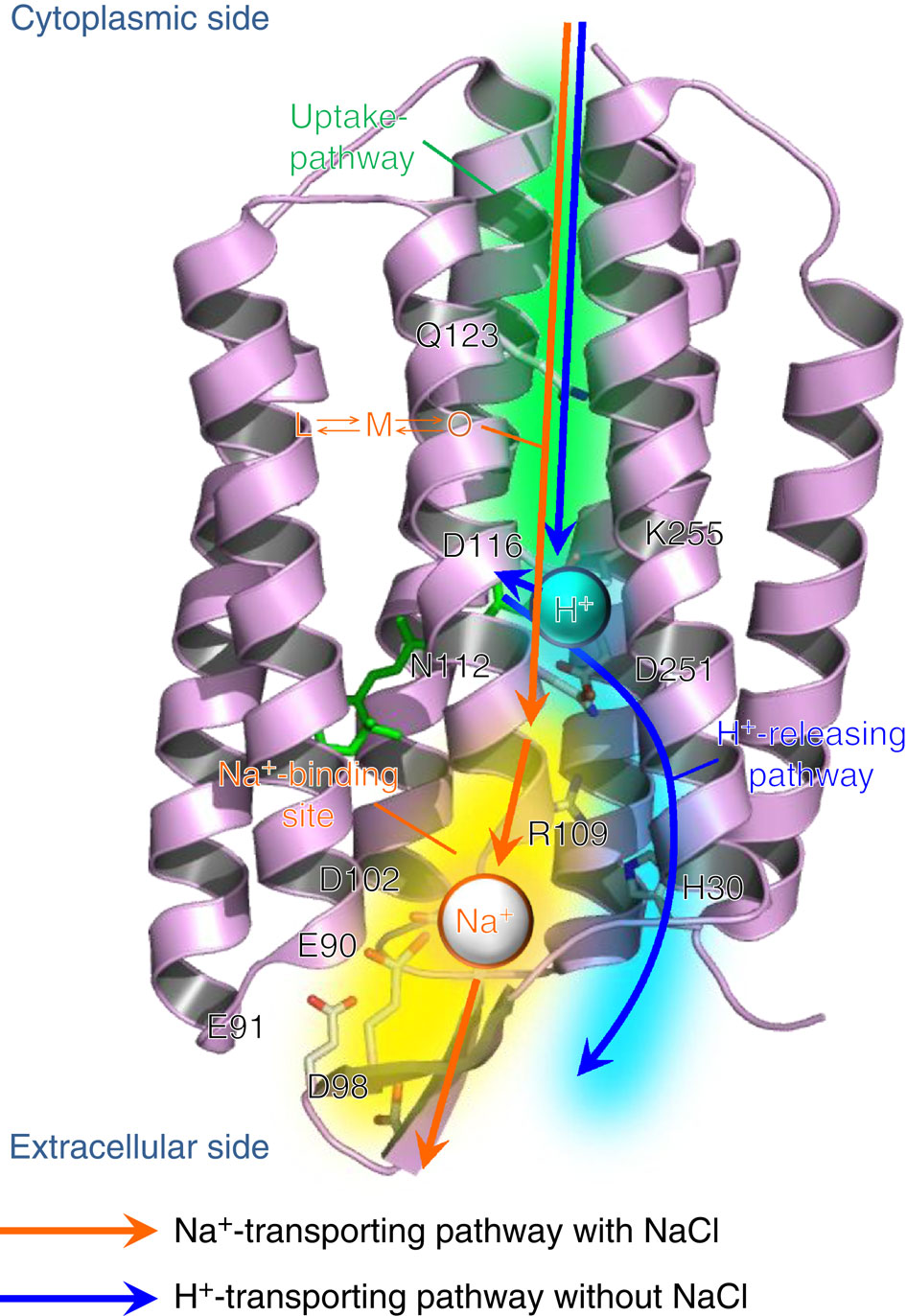
| - | + | KR2 Rhodopsin sodium pump, viewed side-on, showing ion movement and selected molecular interactions | |
| - | + | [[File:RhodopsinKR2.png|400px|center]] | |
| - | + | Credit: Keiichi Inoue et. al (Nature 2013) | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 02:37, 28 September 2013
ABSTRACT
Fresh water shortages affect half of the
world’s population (currently 7 billion persons). In the next 25 years the
numbers of people impacted by severe water shortages is expected to
increase four fold. Shortage of potable water has been estimated to account for
80-90% of disease and 30% of mortality for humanity. Roughly
half of all accessible fresh waters (rivers, lakes and underground aquifers)
are estimated to be in use by the current world population. Given only 2.5
percent of the total volume of water is fresh water and this percentage is
shrinking with global warming, fresh water is becoming a critical limiting
resource for human populations. The goal of this project is to develop an
efficient, green, scalable, low cost solution for desalinization of sea water and brackish water. Our solution is creation of a biofilm (biomachine) which uses sunlight to pump sodium chloride across the biofilm for the desalinization of salt and brackish waters, essentially replacing the pump, membrane, and energy source of a reverse osmosis machine.
The project also provides a scaffold for possible biofilm
cleanup of heavy metals and other toxic ionic pollutants.
Project Goals
1. Identify a prototype polarizable
prokaryotic biofilm species
2. Identify and Clone protein tagging peptides
for intracellular targeting of pumps and channels to specific cellular poles.
3. Identify and Clone a series of sunlight driven ion pumps (halorhodopsins)
4. Identify and Clone a series of ion channels
5. Assemble appropriate tag-pump/channel constructs for assessing NaCl movement across the biofilm.
Experimental Approach
1.
Identify a prototype polarizable
prokaryotic biofilm species. Our
project aims to create a microbial desalination system using the bacteria Caulobacter crescentus. Caulobacter crescentus
was chosen in our initial experiments for five reasons- 1) it is non-pathogenic
and widely distributed in the fresh water aquatic environment; 2) it is
polarized, making it possible to orient ion pumps and channels on specific sides
(poles) of the bacteria; 3) the stalk allows Caulobacter to attach itself to
solid surfaces where it forms a monolayer of cells (biofilm);
4) a set of plasmids is available for transformation of Caulobacter; and 5) experimental fluorescent
tagged protein models are available for investigation of protein tagging
mechanisms of pole formation (stalk and flagellum) in Caulobacter. Please see the
following key references-
Transcriptional Profiling of Caulobacter crescentus during
Growth on Complex and Minimal Media. Hottes et al. Journal of Bacteriology 186(5):1448-1461(2004).
2.
Identify and Clone protein tagging peptides
for intracellular targeting of pumps and channels to specific cellular poles. A
series of protein tags have been identified in pole formation in Caulobacter.
These include DivJ, TipF, StpX, and PflI. Cloning of these
tags are currently underway. Please see the following key references-
PflI, a Protein Involved in Flagellar
Positioning in Caulobacter. Obuchowski and Jacobs-Wagner. Journal of Bacteriology 190(5):1718-1729 (2008).
Protein Sequences and Cellular Factors Required
for Polar Localization of Histidine Kinase in Caulobacter crescentus. Sciochetti et
al., Journal of Bacteriology 184(21):6037-6049 (2002).
Protein localization and dymanics
within a bacterial organelle. Hughes et al.
PNAS:107(12):559-5604(2010).
3.
Identify and Clone a series of sunlight driven ion pumps (halorhodopsins). Candidate channels
include the halorhodopsins and bacterorhodopsins.
We have cloned one new Na pumping halorhodopsin, KR2,
a protein from a marine flavobacterium, Krokinobacter eikastus. Please
see the following key reference- A light-driven sodium
ion pump in marine bacteria. Inoue et al. Nature Communications DOI: 10.1038/ncomms2689
April 9, 2013. We are investigating function of BBaK559000 a BioBrick containing a halorhodopsin.
We are also investigating other sources of halorhodopsins.
4.
Identify and Clone a series of ion channels. We have not yet cloned any ion channels. These proteins are our next
goal for gene cloning.
5.
Assemble appropriate tag-pump/channel constructs for assessing NaCl movement across the biofilm.
Work is in progress for creation of
transforming plasmids coding for fusion proteins which
include the targeting peptide, the pump or channel and a fluorescent tag. These
plasmids will be used to transform Caulobacter. Function will be assessed by determining sub-cellular localization
of the fluorescent tag and assessment for sodium and chloride flux across the biofilm.
For Experimental Results,
see:
Project
Cell polarization
Ion Pumps
BioFilm
Credit: Michael Kolbe et. al (Science, 2000) http://opm.phar.umich.edu/
KR2 Rhodopsin sodium pump, viewed side-on, showing ion movement and selected molecular interactions
Credit: Keiichi Inoue et. al (Nature 2013)
 "
"