Team:NTU-Taida/Notebook/Journal/August
From 2013.igem.org
(→Check) |
|||
| (8 intermediate revisions not shown) | |||
| Line 25: | Line 25: | ||
===Get plasmids:=== | ===Get plasmids:=== | ||
<html> | <html> | ||
| - | <table> | + | <table class="tableizer-table"> |
<tr class="tableizer-firstrow"><th>gene</th><th>name</th><th>conc(ng/ul)</th></tr> | <tr class="tableizer-firstrow"><th>gene</th><th>name</th><th>conc(ng/ul)</th></tr> | ||
<tr><td>Pc-RBS-cinR-tt</td><td>Pc Acin-2</td><td>144.3</td></tr> | <tr><td>Pc-RBS-cinR-tt</td><td>Pc Acin-2</td><td>144.3</td></tr> | ||
| Line 47: | Line 47: | ||
</html> | </html> | ||
<html><a name="805"></a></html> | <html><a name="805"></a></html> | ||
| + | |||
==8/5== | ==8/5== | ||
===Digestion: (biobrick part: restriction enzyme) for 2 hours=== | ===Digestion: (biobrick part: restriction enzyme) for 2 hours=== | ||
| Line 107: | Line 108: | ||
==8/6== | ==8/6== | ||
===Transformation results:=== | ===Transformation results:=== | ||
| - | + | All plates grew. Inoculation of the followings: (5 tubes each, 2 tubes fail, with a total of 78 tubes) | |
#pCI-E1 | #pCI-E1 | ||
#pCI-B5 | #pCI-B5 | ||
| Line 124: | Line 125: | ||
#I13522 | #I13522 | ||
#I13521 | #I13521 | ||
| - | #S03335 | + | #S03335 |
===Original biobrick PCR results are similar to yesterday’s.=== | ===Original biobrick PCR results are similar to yesterday’s.=== | ||
| Line 145: | Line 146: | ||
==8.8== | ==8.8== | ||
===Sequencing results:=== | ===Sequencing results:=== | ||
| - | + | IV. 130806102 | |
#Pc Acin-5 F | #Pc Acin-5 F | ||
#Bcin Cg-1 F | #Bcin Cg-1 F | ||
| Line 160: | Line 161: | ||
#RBS-CI-tt | #RBS-CI-tt | ||
Results: PcAcin/ Bcin are wrong | Results: PcAcin/ Bcin are wrong | ||
| - | |||
===Plasmid extraction=== | ===Plasmid extraction=== | ||
<html> | <html> | ||
| - | <table class> | + | <table class="tableizer-table"> |
<tr class="tableizer-firstrow"><th>Gene</th><th>Name</th><th>size </th><th>conc</th></tr> | <tr class="tableizer-firstrow"><th>Gene</th><th>Name</th><th>size </th><th>conc</th></tr> | ||
<tr><td>pCI-RBS-GFP-tt</td><td>pCI E1 2</td><td>916</td><td>81.5</td></tr> | <tr><td>pCI-RBS-GFP-tt</td><td>pCI E1 2</td><td>916</td><td>81.5</td></tr> | ||
| Line 197: | Line 197: | ||
</html> | </html> | ||
<html><a name="809"></a></html> | <html><a name="809"></a></html> | ||
| + | |||
==8.9== | ==8.9== | ||
===Cloning =Digest+ligase+transform=== | ===Cloning =Digest+ligase+transform=== | ||
| Line 206: | Line 207: | ||
#RBS-LuxR 1 / tt A Lux | #RBS-LuxR 1 / tt A Lux | ||
#PcA C1-3 / Cg,Cm,Cb,Cl ABC Rhl g,m,b,l | #PcA C1-3 / Cg,Cm,Cb,Cl ABC Rhl g,m,b,l | ||
| - | #PcA / C1 | + | #PcA / C1 PcA C1(or PcA rhl) |
Because some bands are irregular, these bands fail to be cloned. | Because some bands are irregular, these bands fail to be cloned. | ||
| Line 264: | Line 265: | ||
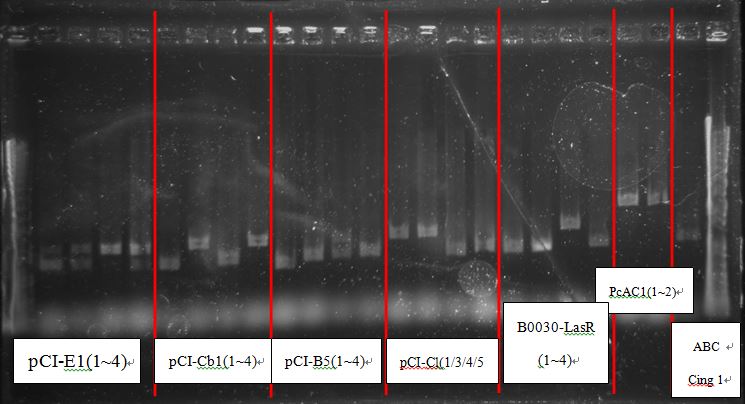
[[File:NTU-Taida-journal-August-8.jpg|700px|thumb|center]] | [[File:NTU-Taida-journal-August-8.jpg|700px|thumb|center]] | ||
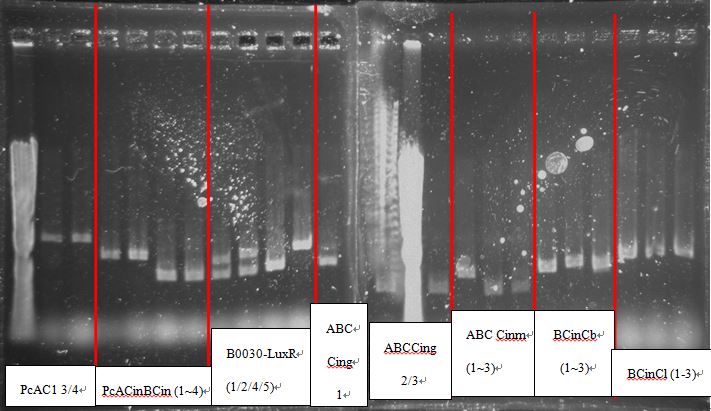
[[File:NTU-Taida-journal-August-9.jpg|700px|thumb|center]] | [[File:NTU-Taida-journal-August-9.jpg|700px|thumb|center]] | ||
| - | + | PcAcin bands are strange! | |
===Plasmid miniprep=== | ===Plasmid miniprep=== | ||
| Line 330: | Line 331: | ||
#PcD: S/P | #PcD: S/P | ||
#BLas 3: X/P | #BLas 3: X/P | ||
| - | + | →RBS-CinR didn’t had insert. Probably failed? | |
===Ligation=== | ===Ligation=== | ||
| Line 367: | Line 368: | ||
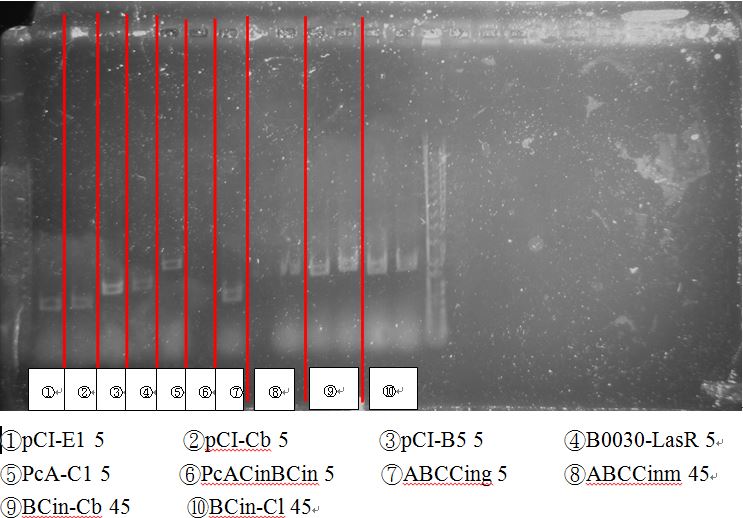
[[File:NTU-Taida-journal-August-12.jpg|700px|thumb|center]] | [[File:NTU-Taida-journal-August-12.jpg|700px|thumb|center]] | ||
[[File:NTU-Taida-journal-August-13.jpg|700px|thumb|center]] | [[File:NTU-Taida-journal-August-13.jpg|700px|thumb|center]] | ||
| - | + | BCI-pCI B5 looked too short | |
===Plasmid miniprep=== | ===Plasmid miniprep=== | ||
Latest revision as of 21:25, 27 September 2013
August
8/1
Inoculation & incubation
- Pc+AcinR (5 tubes)
- BcinR+CGFPmut(E1) (5 tubes)
- B0030-mTagBFP+B0015 (5 tubes)
- B0030-Luci+B0015 (5 tubes)
- B0030-CI+B0015 (5 tubes)
8/2
Check the following tubes:
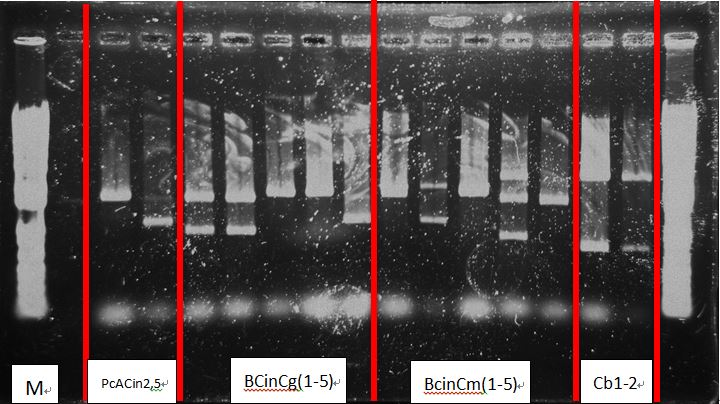
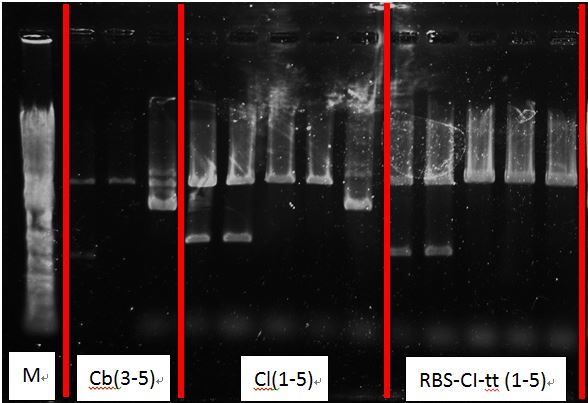
- PcAcin2,5
- BcinCg1-5
- BcinCm1-5
- Cb1-5
- Cl 1-5
- RBS-CI-tt 1-5
Get plasmids:
| gene | name | conc(ng/ul) |
|---|---|---|
| Pc-RBS-cinR-tt | Pc Acin-2 | 144.3 |
| Pc Acin-5 | 142.6 | |
| Pcin-RBS-cinR-RBS-GFPmut-tt | Bcin Cg-1 | 182.9 |
| Bcin Cg-2 | 173.9 | |
| Bcin Cg-3 | 120 | |
| Bcin Cg-4 | 129.7 | |
| Pcin-RBS-cinR-RBS-mCherryt-tt | Bcin Cm-1 | 104.4 |
| Bcin Cm-2 | 105.4 | |
| Bcin Cm-3 | 103.2 | |
| Bcin Cm-4 | 118.5 | |
| RBS-BFP-tt | Cb-1 | 186.8 |
| Cb-2 | 191.9 | |
| Cb-3 | 214.3 | |
| RBS-Luciferase-tt | Cl-1 | 266.4 |
| Cl-2 | 249.1 | |
| RBS-CI-tt | RBS-CI-tt-1 | 159.3 |
| RBS-CI-tt-2 | 166.1 |
8/5
Digestion: (biobrick part: restriction enzyme) for 2 hours
- pCI: S/P
- E1(B0030-GFPmut3-tt): X/P
- B5(B0030-mCherry-tt): X/P
- Cb3(B0030-mTagBFP-tt): X/P
- Cl1(B0030-luciferase-tt): X/P
- PcA(pConst-B0030-RhlR-tt): S/P
- C1(pRhl-B0030-RhlR): X/P
- B0030: S/P
- LasR: X/P
- LuxR: X/P
- PcACin2 & 5:S/P
- BCin3:X/P, S/P
- BCinCm(pCin-RBS-CinR-RBS-mCherry-tt): X/P
- BCinCg(pCin-RBS-CinR-RBS-GFP-tt): X/P
Ligtion(gene part-gene part)
- pCI-E1
- pCI-B5
- pCI-Cb
- pCI-Cl
- B0030-LasR
- B0030-LuxR
- PcA-C1
- PcACin-BCin x2
- PcACin-BCinCm x2
- PcACin-BCinCg x2
- BCin-Cb
- BCin-Cl
Transform total 20 plates
Ampicillin resistant plates (total of 15 plates)
- pCI-E1
- pCI-B5
- pCI-Cb
- pCI-Cl
- B0030-LasR
- B0030-LuxR
- PcA-C1
- PcACin-BCin x2
- PcACin-BCinCm x2
- PcACin-BCinCg x2
- BCin-Cb
- BCin-Cl
Chloramphenicol resistant plates (total of 5 plates)
- K575033
- K575024
- I13522
- I13521
- S03335
PCR of original biobrick
Band length is strange. Needs further PCR.
Functional assay testing ~No results
8/6
Transformation results:
All plates grew. Inoculation of the followings: (5 tubes each, 2 tubes fail, with a total of 78 tubes)
- pCI-E1
- pCI-B5
- pCI-Cb
- pCI-Cl
- B0030-LasR
- B0030-LuxR
- PcA-C1
- PcACin-BCin x2
- PcACin-BCinCm x2
- PcACin-BCinCg x2
- BCin-Cb
- BCin-Cl
- K575033
- K575024
- I13522
- I13521
- S03335
Original biobrick PCR results are similar to yesterday’s.
Functional assay test~ No results
8.7
Trouble shooting: Used the wrong primer for original biobrick PCR
New PCR results (annealing temperature at 52∘C)
Further process:
- PCR with rTaq polymerase: add 1 A to terminals
- Ligation the three tubes overnight (plasmid undergo blue-white selection)
Check results:
8.8
Sequencing results:
IV. 130806102
- Pc Acin-5 F
- Bcin Cg-1 F
- Bcin Cg-1 R
- Bcin Cg-3 F
- Bcin Cg-3 R
- Bcin Cm-1 F
- Bcin Cm-1 R
- Bcin Cm-2 F
- Bcin Cm-2 R
- Cb-1 F
- Cb-1 R
- Cl-1
- RBS-CI-tt
Results: PcAcin/ Bcin are wrong
Plasmid extraction
| Gene | Name | size | conc |
|---|---|---|---|
| pCI-RBS-GFP-tt | pCI E1 2 | 916 | 81.5 |
| pCI E1 4 | 74.1 | ||
| pCI-RBS-BFP-tt | pCI Cb 2 | 898 | 97.4 |
| pCI Cb 4 | 110 | ||
| pCI-RBS-mCherry-tt | pCI B5 2 | 907 | 102.1 |
| pCI B5 3 | 101.3 | ||
| pCI-RBS-luciferase-tt | pCI Cl 4 | 1129 | 85.4 |
| pCI Cl 5 | 146.6 | ||
| RBS-LasR | RBS lasR 1 | 867 | 128.7 |
| RBS lasR 2 | 125.7 | ||
| Pc-RBS-RhlR-tt-pRhl-RBS-RhlR | PcA C1 3 | 1705 | 93 |
| PcA C1 4 | 74.5 | ||
| Pc-RBS-CinR-tt-pCin-RBS-CinR | PcAcin Bcin 1993 | wrong sequence | |
| RBS-LuxR | RBS luxR 1 796 69.1 | ||
| RBS luxR 2 | 101.4 | ||
| Pc-RBS-CinR-tt-pCin-RBS-CinR-RBS- | |||
| GFP-tt | ABC cing | 2860 | |
| Pc-RBS-CinR-tt-pCin-RBS-CinR-RBS | |||
| -mCherry-tt | ABC cinm | 2851 | |
| pCin-RBS-CinR-RBS-BFP-tt | Bcin Cb 1 | 1876 | 118.2 |
| Bcin Cb 2 | 113 | ||
| pCin-RBS-CinR-RBS-Luciferas-tt | Bcin Cl 1 | 2110 | 141.3 |
| Bcin Cl 2 | 87.1 | ||
| K575033 | 214.6 | ||
| K575024 | 331.9 | ||
| I13522 | 236.6 | ||
| I13521 | 380 | ||
| S03355 | 274.5 |
8.9
Cloning =Digest+ligase+transform
- Pc(18C) / Acin Pc A Cin
- Pc(18A) / Acin Pc A Cin
- pCinR / RBS-CinR B Cin
- pLuxR / RBS-LuxR 2 B Lux
- pLasR /RBS-LasR 1 B Las
- RBS-LuxR 1 / tt A Lux
- PcA C1-3 / Cg,Cm,Cb,Cl ABC Rhl g,m,b,l
- PcA / C1 PcA C1(or PcA rhl)
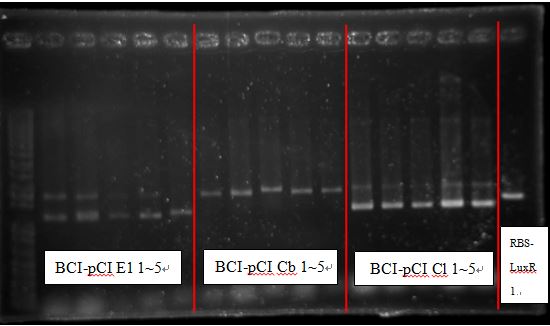
Because some bands are irregular, these bands fail to be cloned.
PCR product has undergone blue-white selection, and will need to be incubated under Kanamycin and Ampicillin LB broth.
Functional assay
Run the ELISA plate reader with the control group which keeps expressing GFP. Still no successful results yet.
8.12
Digestion(gene part : restriction enzyme)
- B0030-CI-tt(=BCI) 2: S/P
- pCI-E1(GFP) 2: X/P
- pCI-B5(mCherry) 3: X/P
- pCI-Cb(BFP) 4: X/P
- pCI-Cl(luci) 5: X/P
- LuxR: X/P
Ligation (some material were from past digestion products, which are already well cut)
- BCI-pCI E1
- BCI-pCI B5
- BCI-pCI Cb
- BCI-pCI Cl
- B0030-LuxR
- pCin-[B0030-CinR]
Transformation: 6 plates (Ampicillin resistant)
- BCI-pCI E1
- BCI-pCI B5
- BCI-pCI Cb
- BCI-pCI Cl
- B0030-LuxR
- pCin-[B0030-CinR]
Inoculation (5 tubes for each plate, total of 20 tubes)
- Pc-ACin (18C) Amp resistant
- Pc-ACin (18A) Amp resistant
- BLas(pLas-B0030-LasR) Amp resistant
- PcA-C1 Amp resistant
8.13
Transform results(All grew, except BCI-pCI Cb grew too much )
- BCI-pCI E1
- BCI-pCI B5
- BCI-pCI Cb (grow too much, lack single colony)
- BCI-pCI Cl
- B0030-LuxR
- pCin-[B0030-CinR]
Inoculation/ Incubation(Ampicillin resistant, each plate inoculates 5 tubes, with a total of 25 tubes)
- BCI-pCI E1
- BCI-pCI B5
- BCI-pCI Cl
- B0030-LuxR
- pCin-[B0030-CinR] (=BCin)
Check
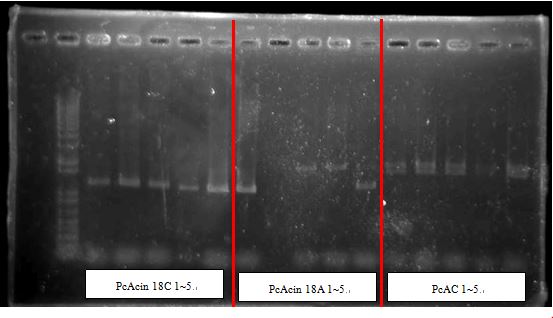
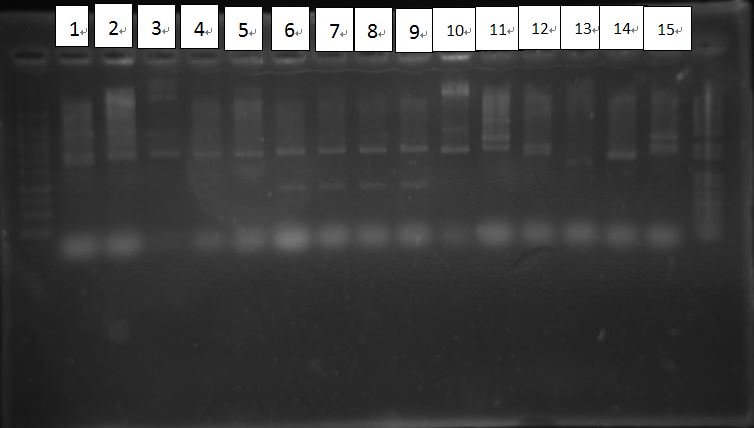
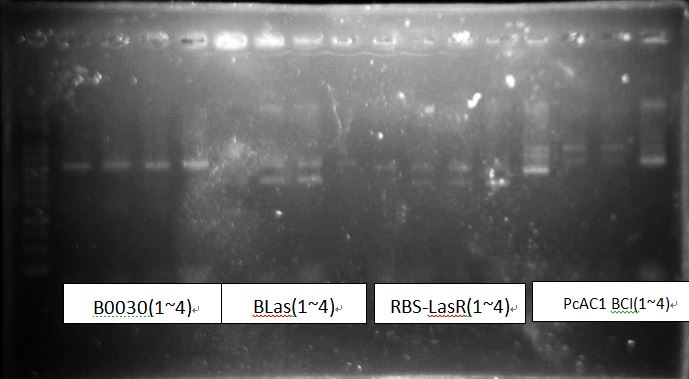
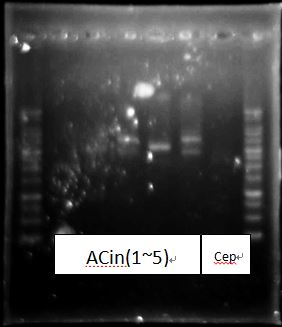
PcAcin bands are strange!
Plasmid miniprep
- BLas 1,2
- PcACin 18C 1,2
- PcACin 18A 2,5
- PcAC1 1,2
Digestion (gene part: restriction enzyme)
- pLas: S/P
- pRhl: S/P
- pCin 2: S/P
- B5 (mCherry): X/P
- pCI Cb: X/P
Gel electrophoresis: pCI Cb is possible to fail because insert is nowhere seen
Ligation
- pLas-Cb
- pRhl-B5
- pCin-E1
- pCI-Cb (redid it again)
- BCI-pCI B5
- BCI(vector only): to see whether digestion was clean enough.
Transformation (Ampicillin resistant)
- pLas-Cb
- pRhl-B5
- pCin-E1
- pCI-Cb
- BCI-pCI B5
- BCI
8.14
Transform results:
- pCI-Cb didn’t growth
- BCI (vector only) growth a little, prove that digestion was successful
Inoculation (Ampicillin resistant, 5 tubes each plus 1 tube of ACE)
- pLas-Cb
- pRhl-B5
- pCin-E1
- BCI-pCI B5
- ACE : for stock
Original biobrick
Inoculation 20 tubes (plate 163,134 – promoter PQS) Ampicillin resistant
Check
BCI-pCI Cb looked failed, because inserts of BCIs all looked too short?
RBS-LuxR had strange length~
Plasmid Miniprep
- BCI-pCI E1 1,2
- BCI-pCI Cl 4,5
- BCin 3,4
Digestion
- Pc18C (J23100): S/P
- Pc (J23119): S/P
- RBS-CinR: E/S
- PcAC1 1: S/P
- PcD: S/P
- BLas 3: X/P
→RBS-CinR didn’t had insert. Probably failed?
Ligation
- J23119(Pc18A)-ACin
- PcAC1-B5
- PcAC1-Cb
- PcAC1-Cl
- PcD-BLas
Transformation
- Pc18A-ACin Amp
- PcAC1-B5(ABC Rhl m) Amp
- PcAC1-Cb(ABC Rhl b) Amp
- PcAC1-Cl(ABC Rhl l) Amp
- PcD-BLas Amp
- LuxR Amp: from official gel stab
8.15
Transform Result
- All plates grew~
- Pc(18A)ACin growth(but only a little)! Yet its resistance was Chl…
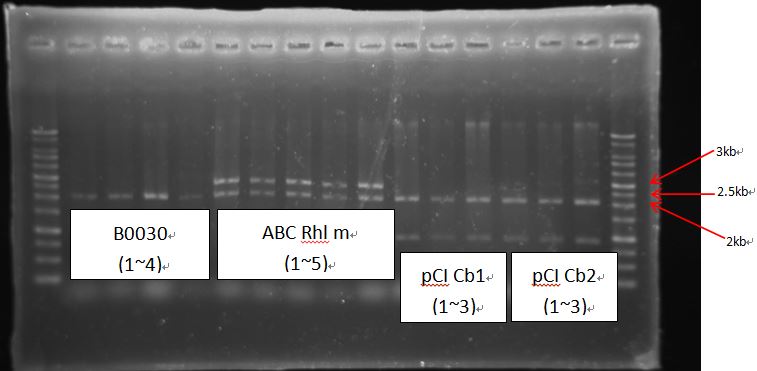
- ABC Rhl m growth too much.
Inoculation (Ampicillin resistant, 22 tubes total)
- ABC Rhl b
- ABC Rhl l
- PcD-BLas
- LuxR
- Pc(18A)-ACin: check resistance, 2 tubes
Original biobrick
- Inoculation 20 tubes (plate 159,130 - pqsR) Amp
- Check result (Ppqs)
Check
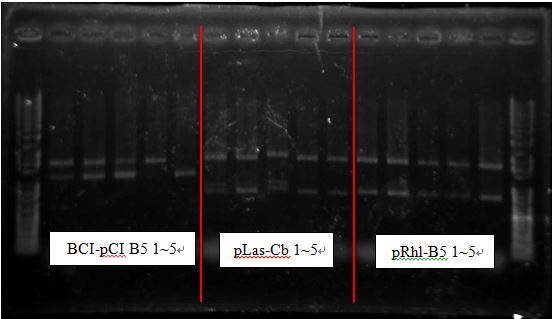
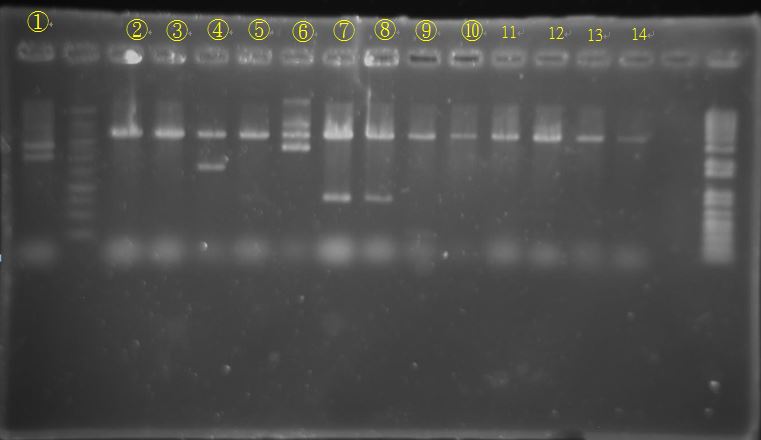
BCI-pCI B5 looked too short
Plasmid miniprep
- pCin E1 1,4
- BCI-pCI B5 2,3
- pLas-Cb 2,4
- pRhl 1,5
Digestion
- pCI 1: S/P
- Cb 1: X/P
- Cb 2: X/P
- Cb 3: X/P
Three Cb looked alike
Ligation
- pCI-Cb1
- pCI-Cb2
- pCI-Cb3
- Pc(18A)-ACin
Transformation
- pCI-Cb1 Amp
- pCI-Cb2 Amp
- pCI-Cb3 Amp
- Pc(18A)-ACin Chl
- ABC Rhl m Amp: transfer to another plate
8.16
Check inoculated bacteria
1: PcD Blas4 2: PcD Blas5 3: PcD Blas3 4: PcD Blas1
5: LuxR 1 6: LuxR 2 7: LuxR 3 8: LuxR 4
9: LuxR 5 10: Rhlb2 11: Rhlb5 12: PcAcin try1
13: PcAcin try2 14: Rhlb1 15: Rhlb2
1: Rhl b4 2: PQSR1 3: PQSR2 4: PQSR3
5: PQSR5 6: PQSR8 7: PQSR10 8: PQSR11
9: PQSR13 10: PQSR14 11: PQSR15 12: PQSR16
13: PQSR18 14: PQSR20
Transform results:
- ABC Rhlm(0)
- pCI-Cb1(0)
- pCI-Cb3(0)
- pCI-Cb2(0)
- Chl-PcACin(18A) (X)
8.19
Plasmid miniprep(genepart-concenration)
PcAC1-Cl 2 170.5 PcAC1-Cl 4 202.9 PcAC1-Cb 1 137.5 PcAC1-Cb 3 140 PcD-Blas 1 148 PcD-Blas 3 159.2 PcD-Blas 4 152.9 PQSR10 373.6 PQSR11 347.4 LuxR2 89.8 LuxR3 101.1
Check ABC-Rhl l
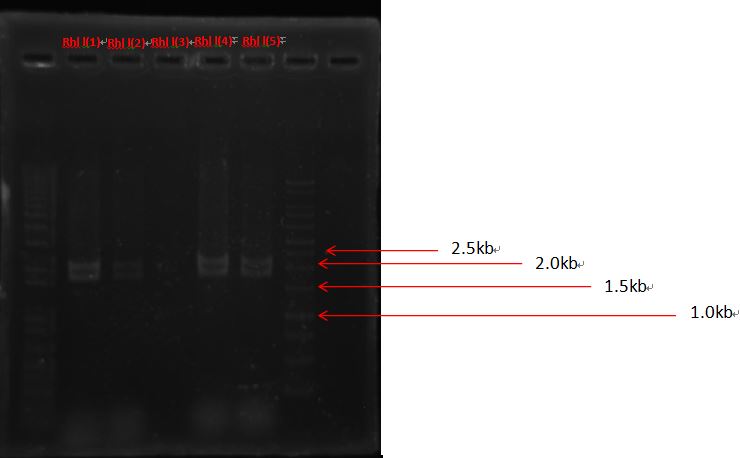
The bands seemed to be weird. It should be around 2.8kb instead of 2k, therefore needs further sequencing to identify.
Digestion
- RBS-CinR: E/S
- pLas Cb#: X/P
- pRhl B5#: X/P
- BCI#: X/P
- RBS-LasR#: E/S→used wrong enzyme
- RBS-LasR#: E/S→used wrong enzyme
- pPQS(19): E/P
- linear pSB1C3: E/P→No results, probably too low concentration
Ligation: some used previous digestion products
- [RBS-CinR]-tt (=ACin)
- pLas-[RBS-LasR] (=BLas)
- PcD-pLas Cb (=OLasCb)
- PcA-pRhl B5 (=ORhlB5)
- PcAC1-BCI
- RBS-LasR
Transformation: total 5 plates(Ampicillin resistant)
- ACin
- BLas
- OLasCb
- ORhlB5
- PcAC1-BCI
- RBS-LasR
Inoculation(Ampicillin resistant, 24 tubes total)
- B0015: 5 tubes
- B0030: 5 tubes
- ABC Rhlm: 5 tubes
- pCI Cb###: each 3 tubes
8.20
Check:
B0015 did not grow at all
Plasmid miniprep:
- B0030-2
- B0030-3
- ABC Rhl m-1
- ABC Rhl m-2
- pCI Cb2-1
- pCI Cb2-2
Transform(1 plate)
pSB1C3 backbone and promoter PQS are transformed after ligation.
Inoculation:(Ampicillin resistant)
- ACin Amp(5 tubes)
- BLas Amp(5 tubes)
- PcAC1-BCI Amp(5 tubes)
- RBS-LasR Amp(5 tubes)
- B0015 Amp(5 tubes)
Sequencing: sending 10 samples(R=reverse primer, F=forward primer)
- BCin# R
- pCI B5 F
- pCI E1 F
- pCI Cl F
- pCin E1 F
- pRhl B5 F
- ABC Rhl l F
- ABC Rhl l R
- ABC Rhl b F
- ABC Rhl b R
8.22
Check:
Plasmid miniprep
- B0015-2,3
- BLas-2,3
- RBS-LasR-3,4
- pCAC1-BCI-2,3
- Acin 1
Digest(gene part-restriction enzyme)
- pPQS19 E/S
- E1-1 E/X
- B5-1 E/X
- Cb3 E/X
- Cl1 E/X
- PcDBLas 3 S/P
Ligation & Transform:
- pPQS- E1 Amp
- pPQS- B5 Amp
- pPQS-Cb Amp
- pPQS-Cl Amp
Inoculation:
- pPQS+pSC1C3 backbone-5 tubes Chloramphenical resistant
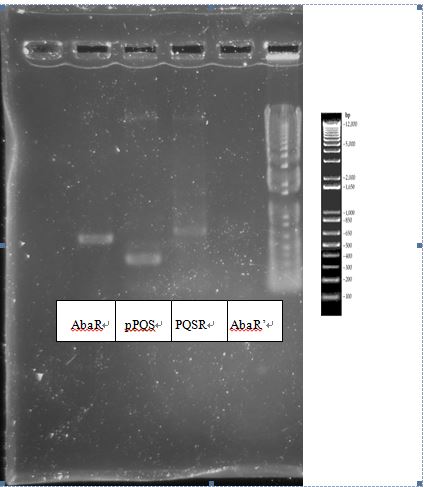
- AbaR (on T vector)-5 tubes Ampicillin+Kanamycin resistant
 "
"