Team:SCUT/Project/Odorant sensing
From 2013.igem.org
Junjiezhang (Talk | contribs) |
Yanzw scut (Talk | contribs) |
||
| (37 intermediate revisions not shown) | |||
| Line 14: | Line 14: | ||
<div id="label"> | <div id="label"> | ||
<div id="mark1"class="labelmenu" style="background-color:#d6ee5a;"> | <div id="mark1"class="labelmenu" style="background-color:#d6ee5a;"> | ||
| - | <p onclick="scroll_1()"> | + | <p onclick="scroll_1()"style="letter-spacing:-1px;">Introduce..</p> |
</div> | </div> | ||
<div id="mark2"class="labelmenu" style="background-color:#cbf4bb;"> | <div id="mark2"class="labelmenu" style="background-color:#cbf4bb;"> | ||
| - | <p onclick="scroll_2()"> | + | <p onclick="scroll_2()"style="padding:2px 0;">Pathway overview</p> |
</div> | </div> | ||
<div id="mark3"class="labelmenu" style="background-color:#efd9cb;"> | <div id="mark3"class="labelmenu" style="background-color:#efd9cb;"> | ||
| - | <p onclick="scroll_3()"> | + | <p onclick="scroll_3()"style="letter-spacing:-1px;">Reconstruct</p> |
</div> | </div> | ||
<div id="mark4"class="labelmenu" style="background-color:#f979a4;"> | <div id="mark4"class="labelmenu" style="background-color:#f979a4;"> | ||
| - | <p onclick="scroll_4()"> | + | <p onclick="scroll_4()">Optimize</p> |
</div> | </div> | ||
<div id="mark5"class="labelmenu" style="background-color:#d3c902;"> | <div id="mark5"class="labelmenu" style="background-color:#d3c902;"> | ||
<p onclick="scroll_5()">Result</p> | <p onclick="scroll_5()">Result</p> | ||
</div> | </div> | ||
| - | <div id=" | + | <div id="mark6"class="labelmenu" style="background-color:#F5F978;"> |
| - | <p onclick=" | + | <p onclick="scroll_6()">Reference</p> |
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 57: | Line 54: | ||
To express exogenous GPCR,our plasmid manipulation is performed in E.coli strain top10. Odr-10 CDS sequence, which is optimized in GenScript, was inserted into the YEP352 shuttle vector with GAL1 promoter and ADH1 terminator.<br><br> | To express exogenous GPCR,our plasmid manipulation is performed in E.coli strain top10. Odr-10 CDS sequence, which is optimized in GenScript, was inserted into the YEP352 shuttle vector with GAL1 promoter and ADH1 terminator.<br><br> | ||
However the odr-10 fails to contain the N-terminal import sequence, which results in the difficulty of localization on plasma membrane when express odr-10 in heterogeneous cells. The overexpressed and inefficient folding odr-10 will stimulate the endoplasmic reticulum stress (ERS) and autophagy pathway. To solve the difficulty of odr-10 localization on the plasma membrane, we try to insert the sequence of rhodopsin signal peptide (rho) into the upstream of odr-10 CDS. Our rho sequence has also been optimized. Additionally, to detect whether our odr-10 has been transcribed and located on the plasma membrane or not, we insert the sequence of GPF in downstream of odr-10 and the sequence of flag-tagged into the position between our rho and odr-10 respectively.<br><br> | However the odr-10 fails to contain the N-terminal import sequence, which results in the difficulty of localization on plasma membrane when express odr-10 in heterogeneous cells. The overexpressed and inefficient folding odr-10 will stimulate the endoplasmic reticulum stress (ERS) and autophagy pathway. To solve the difficulty of odr-10 localization on the plasma membrane, we try to insert the sequence of rhodopsin signal peptide (rho) into the upstream of odr-10 CDS. Our rho sequence has also been optimized. Additionally, to detect whether our odr-10 has been transcribed and located on the plasma membrane or not, we insert the sequence of GPF in downstream of odr-10 and the sequence of flag-tagged into the position between our rho and odr-10 respectively.<br><br> | ||
| - | In the endogenous MAP-kinase pathway in S.cerevisiae, third messenger will stimulate fus1 promoter, which results in mating of the yeast. In like manner, we create the signal reporter device as “fus1+ | + | In the endogenous MAP-kinase pathway in S.cerevisiae, third messenger will stimulate fus1 promoter, which results in mating of the yeast. In like manner, we create the signal reporter device as “fus1+BFP+ADH1 terminator”. However the sequence of fus1 contains Pst1 restrict enzyme site, we use the method of overlap extension to achieve the purpose of site-directed mutagenesis ("CTGCAG" to "CTGGAG"). |
</p> | </p> | ||
</div> | </div> | ||
| Line 80: | Line 77: | ||
<div id="tblock5"> | <div id="tblock5"> | ||
| - | <h1> | + | <h1>Results</h1> |
<p class="text">We delete the FAR1 gene successfully as shown in the figure 4. | <p class="text">We delete the FAR1 gene successfully as shown in the figure 4. | ||
</p> | </p> | ||
<div class="imgC"> | <div class="imgC"> | ||
| - | <img style="width: | + | <img style="width:600px;margin-right:10px;"src="https://static.igem.org/mediawiki/parts/a/a9/Scut_sensing4.png"/> |
<p class="tag"style="width:660px;"><strong>Figure 4: An ethidium-biomide-stained agarose gel showing results of FAR1 gene knockout.</strong><br><br> | <p class="tag"style="width:660px;"><strong>Figure 4: An ethidium-biomide-stained agarose gel showing results of FAR1 gene knockout.</strong><br><br> | ||
<strong>a: </strong>The first lane is a 250bp DNA marker ladder. The lanes 2-5 are products of PCR using p3 and p4 as the primers using the knockout Far1 yeast genome as the template but the lane 6 uses the original yeast genome as the template. The bands are correct size of zeocin which is not in the original S.cerevisiae BY4741.<br> | <strong>a: </strong>The first lane is a 250bp DNA marker ladder. The lanes 2-5 are products of PCR using p3 and p4 as the primers using the knockout Far1 yeast genome as the template but the lane 6 uses the original yeast genome as the template. The bands are correct size of zeocin which is not in the original S.cerevisiae BY4741.<br> | ||
| Line 91: | Line 88: | ||
</div> | </div> | ||
<p class="text">Until now, we have successfully constructed 14 expression plasmids. The sequence of flag-tag, Rho and our olfactory receptor odr-10 has been optimized in genscript.<br> | <p class="text">Until now, we have successfully constructed 14 expression plasmids. The sequence of flag-tag, Rho and our olfactory receptor odr-10 has been optimized in genscript.<br> | ||
| - | <img style="width:700px | + | <img style="width:700px;margin:10px 0;"src="https://static.igem.org/mediawiki/parts/a/ab/Scut_sensing5.png"/><br> |
We have constructed three devices (DⅠ, DⅡ& DⅢ) to express the olfactory receptor(OR).<br> | We have constructed three devices (DⅠ, DⅡ& DⅢ) to express the olfactory receptor(OR).<br> | ||
| - | DⅠ: BBa_K1072013 <img style=" | + | DⅠ: <a href="http://parts.igem.org/Part:BBa_K1072013" class="brick">BBa_K1072013</a> <img style="height:60px;"src="https://static.igem.org/mediawiki/parts/b/b7/Scut_sensing6.jpg"/><br> |
| - | DⅡ: BBa_K1072014 <img style=" | + | DⅡ: <a href="http://parts.igem.org/Part:BBa_K1072014" class="brick">BBa_K1072014</a> <img style="height:60px;"src="https://static.igem.org/mediawiki/parts/a/a3/Scut_sensing7.jpg"/><br> |
| - | DⅢ: BBa_K1072015<img style=" | + | DⅢ: <a href="http://parts.igem.org/Part:BBa_K1072015" class="brick">BBa_K1072015</a> <img style="height:60px;"src="https://static.igem.org/mediawiki/parts/2/2c/Scut_sensing8.jpg"/><br><br> |
| - | And then three odorant sensing systems(SⅠ, SⅡ& SⅢ) were inserted into shuttle plasmid YEplac181 | + | And then three odorant sensing systems(SⅠ, SⅡ& SⅢ) were inserted into shuttle plasmid YEplac181. Reporter device was <a href="http://parts.igem.org/Part:BBa_K1072020" class="brick">BBa_K1072020</a>, pFus1 + BFP + ADH1 terminator.<br> |
| - | + | SⅠ:<a href="http://parts.igem.org/Part:BBa_K1072016" class="brick">BBa_K1072016</a> <img style="height:60px;"src="https://static.igem.org/mediawiki/parts/7/7a/Scut_sensing9.jpg"/><br> | |
| - | SⅡ: BBa_K1072017 <img style=" | + | SⅡ: <a href="http://parts.igem.org/Part:BBa_K1072017" class="brick">BBa_K1072017</a> <img style="height:60px;"src="https://static.igem.org/mediawiki/parts/2/21/Scut_sensing10.jpg"/><br> |
| - | SⅢ: BBa_K1072018 <img style=" | + | SⅢ: <a href="http://parts.igem.org/Part:BBa_K1072018" class="brick">BBa_K1072018</a> <img style="height:60px;"src="https://static.igem.org/mediawiki/parts/0/01/Scut_sensing11.jpg"/><br> |
| - | Our olfactory receptor odr-10 has been transformed into <i>S.cerevisiae</i>. we transformed | + | Our olfactory receptor odr-10 has been transformed into <i>S.cerevisiae</i>. we transformed SⅠ<a href="http://parts.igem.org/Part:BBa_K1072016" class="brick">BBa_K1072016</a> into <i>S.cerevisiae</i> and cultured the cells until they reached the exponential phase(OD600, in the range of 1-2), then we diluted broth to OD600 0.5 and cultured in the selection media containing 2% galactose at 30℃ for 18h.<br> |
Finally, we found that GFP, Flag tag signal and BFP were bright by confocal microscope,which means that Odr-10 gene have be translated, located and induced MAPK pathway successfully, as showing in figure 5.Flag tag was detected around plasma membrane, as we can see in figure 6. | Finally, we found that GFP, Flag tag signal and BFP were bright by confocal microscope,which means that Odr-10 gene have be translated, located and induced MAPK pathway successfully, as showing in figure 5.Flag tag was detected around plasma membrane, as we can see in figure 6. | ||
</p> | </p> | ||
<div class="imgC"> | <div class="imgC"> | ||
| - | <img style="width:600px;margin-right:10px;"src="https://static.igem.org/mediawiki/ | + | <img style="width:600px;margin-right:10px;"src="https://static.igem.org/mediawiki/igem.org/a/aa/Scut_sensing11a.png"/> |
| - | <p class="tag"style="width:660px;"><strong>Figure 5:</strong> | + | <p class="tag"style="width:660px;"><strong>Figure 5:</strong> Transforming Odr-10 with flag tag and reporter gene into yeast,GFP, flag tag signal and BFP was detected successfully, meaning that the whole pathway was stimulated by diacetyl.</p> |
</div> | </div> | ||
<div class="imgC"> | <div class="imgC"> | ||
| - | <img style="width:600px;margin-right:10px;"src="https://static.igem.org/mediawiki/ | + | <img style="width:600px;margin-right:10px;"src="https://static.igem.org/mediawiki/igem.org/0/0c/Scut_sensing12a.jpg"/> |
| - | <p class="tag"style="width:660px;"><strong>Figure 6:</strong> | + | <p class="tag"style="width:660px;"><strong>Figure 6:</strong> FLAG immunolocalization of yeast cells expressing Odr-10 with flag tag, which means that Odr-10 protein locate at plasma membrane successfully.</p> |
</div> | </div> | ||
| - | <p class="text">Additionally, we transformed SⅡ system into <strong>S.cerevisiae</strong> and cultured the cells as above method. We found that SⅡsystem, | + | <p class="text">Additionally, we transformed SⅡ system into <strong>S.cerevisiae</strong> and cultured the cells as above method. We found that SⅡsystem, <a href="http://parts.igem.org/Part:BBa_K1072017" class="brick">BBa_K1072017</a>, expresses bright blue fluorescence under UV field under 400ms exposure time Figure 7. Both BBa_K1072016 and BBa_K1072017 work meaning that Brho isn’t the necessary or don’t work in yeast. Odr-10 successful localization may result from the interior signal peptide of cDNA of Odr-10. |
</p> | </p> | ||
<div class="imgC"> | <div class="imgC"> | ||
| - | <img style="width:600px | + | <img style="width:600px;"src="https://static.igem.org/mediawiki/igem.org/4/43/Scut_sensing13a.jpg"/> |
| - | <p class="tag"><strong>Figure 7.</strong></p> | + | <p class="tag"style="width:580px;"><strong>Figure 7.</strong>Transforming Odr-10 with flag tag and Brho into yeast, BFP brighten,meaning that the whole pathway work and demonstrate that Brho isn't the necessary factors expressing Odr-10 in yeast.</p> |
</div> | </div> | ||
<p class="text"> | <p class="text"> | ||
In order to optimize the pathway and achieve our expectation, we constructed Gpa1-odr3 chimera (chi) device to enhance the intension of signal transduction.<br> | In order to optimize the pathway and achieve our expectation, we constructed Gpa1-odr3 chimera (chi) device to enhance the intension of signal transduction.<br> | ||
| - | The chimera, carboxy-terminal five amino acids of Gpa1 are replaced with corresponding residues from odr-3 subunit, will not only enhance the affinity between Gpa1 with odr-10 from <i>caenorhabditis elegans</i>, but also not influence the affinity between Gpa1 with Gβγ. We utilize the sequence (1000bp) upstream of Gpa1 to serve as the pGpa1 and create a novel biobrick, BBa_K1072011. To make it can be adapted to principle of IGEM, we successfully mutate the restriction sites of EcoR1 and Spe1in the sequence through site-specific mutagenesis.<br><br> | + | The chimera, carboxy-terminal five amino acids of Gpa1 are replaced with corresponding residues from odr-3 subunit, will not only enhance the affinity between Gpa1 with odr-10 from <i>caenorhabditis elegans</i>, but also not influence the affinity between Gpa1 with Gβγ. We utilize the sequence (1000bp) upstream of Gpa1 to serve as the pGpa1 and create a novel biobrick, <a href="http://parts.igem.org/Part:BBa_K1072011" class="brick">BBa_K1072011</a>. To make it can be adapted to principle of IGEM, we successfully mutate the restriction sites of EcoR1 and Spe1in the sequence through site-specific mutagenesis.<br><br> |
| - | The chi device: <img style=" | + | The chi device: <img style="height:60px;"src="https://static.igem.org/mediawiki/igem.org/b/b3/Scut_sensing14a.jpg"/> |
</p> | </p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
Latest revision as of 02:34, 28 September 2013
Introduction
We aim to reconstruct and model the simplest communication in nature, like a flow of information between eukaryotes and prokaryotes through odorant. To accomplish this mission that S.cerecisiae communicate with E.coli, we require to fabricate a kind of dedicated biomimetic nose for our yeast and single out a special and adapted volatile to serve as their “language”.
After a long discussion, argument and selection, finally we decided to select a complex and 7-transmembrane receptors, GPCR, Odr-10, as nose and specific odorant, diacetyl, as the language. In order to construct the system precisely and efficiently, we constructed the Odr-10 expression system and modified the downstream pathway, deleting Gpa1 and Far1 gene then reconstructing and expressing chimeric Gpa1.
Pathway Overview
S.cerevisiae is selected as the protagonist of our system owing to the clear genetic background and its natural advantages to express G protein family. The endogenous receptors, Step2, a G protein coupled receptors (GPCRs), will be activated when they sense a/αpheromone, as description of figure 1.
In yeast, the pheromones are bonds to a GPCR, Sted2, and active downstream pathway. It is composed of Gpa1p (Gα), Ste4p (Gβ) and Ste18p (Gγ) subunits. The GPCR induces the complex of Gα and Gβγ, resulting in the separation of Gα and Gβγ. The Gβγ particle subsequently transmits the signal to a MAP kinase pathway, comprising Ste11p, Ste7p, and Fus3p held in a complex with the Ste5 protein. Fus3p then activates the cyclin-dependent kinase inhibitor Far1p and the transcription factor Ste12p to bring about cell-cycle arrest.
It is worthy to announce that a major negative regulator of the pathway, Sst2p, is a GTPase activating factors that return Gα to its inactive state which generally would be deleted to increase the sensitivity. Finally, a reporter of this pathway was under the control of a Ste12p inducible promoter, Fus1 linking BFP, GFP or HIS3.

Figure 1. Comparison between endogenous GPCR pathway in yeast and motified Odr10 pathway.
Diacetyl is utilized as our "language" for our communication between E.coli and S.cerevisiae. Diacetyl can be released by modified E.coli as a kind of special intermediate in metabolism pathway which was described in Oscillating odorant part. We take advantage of the modeling olfactory receptors (ORs) odr-10, which is from caenorhabditis elegans and specific to diacetyl. Additionally, diacetyl may be harmful to humans when inhaled frequently and diacetyl-induced bronchiolitis obliterans is one of the representative diseases. Consequently, our biomimetic nose can be utilized to detect the diacetyl in our surroundings more sensitively.
Pathway reconstruction
To express exogenous GPCR,our plasmid manipulation is performed in E.coli strain top10. Odr-10 CDS sequence, which is optimized in GenScript, was inserted into the YEP352 shuttle vector with GAL1 promoter and ADH1 terminator.
However the odr-10 fails to contain the N-terminal import sequence, which results in the difficulty of localization on plasma membrane when express odr-10 in heterogeneous cells. The overexpressed and inefficient folding odr-10 will stimulate the endoplasmic reticulum stress (ERS) and autophagy pathway. To solve the difficulty of odr-10 localization on the plasma membrane, we try to insert the sequence of rhodopsin signal peptide (rho) into the upstream of odr-10 CDS. Our rho sequence has also been optimized. Additionally, to detect whether our odr-10 has been transcribed and located on the plasma membrane or not, we insert the sequence of GPF in downstream of odr-10 and the sequence of flag-tagged into the position between our rho and odr-10 respectively.
In the endogenous MAP-kinase pathway in S.cerevisiae, third messenger will stimulate fus1 promoter, which results in mating of the yeast. In like manner, we create the signal reporter device as “fus1+BFP+ADH1 terminator”. However the sequence of fus1 contains Pst1 restrict enzyme site, we use the method of overlap extension to achieve the purpose of site-directed mutagenesis ("CTGCAG" to "CTGGAG").
Pathway optimization
There is no doubt that signal sensing part and signal reporter part are prerequisite, however, things cannot be that simple because the poor affinity between the Gα(Gpa1) and heterogeneous GPCRs will slash the intensity and sensibility of our signal pathway. Similarly, transforming the yeast with full-length odr-3, which is the Gα in the caenorhabditis elegans, also has limited affinity for Gβγ of yeast.
A preferential method is that we can create a chimera of Gpa1/odr-3 and only the carboxy-terminal five amino acids of Gpa1 are replaced with corresponding residues from odr-3subunit. This design is owing to the following two detections: first of all, the principal receptor specificity determinants reside at the very carboxy-terminus of Gαsubunits; secondly, The last eight amino acids of Gα are not resolved on the Gαβγ crystal structures, meaning that the exchanged regions have an undefined conformation extending from the end of the 5α helix Figure 2.

Figure 2. Gprotein heterotrimer. G-proteins are signaling molecules found on the inner side of cell membranes. This structure shows a complete complex of three chains with GDP bound.
In order to achieve the communication between E.coli and S.cerevisiae, we need the heterologous receptor, odr-10, successfully express in the S.cerevisiae to detect the diacetyl produced by E.coli. The functional coupling of GPCRs has been achieved typically by modifications to the G protein alpha subunit, Gpa1p and Gβγ signals to a MAP kinase cascade to induce cell-cycle arrest by expressing FAR1 gene and transcription of mating genes. Therefore, successful coupling of foreign GPCRs need to only express chimeric Gαsubunit by deletion of Gpa1 and disable the cell-cycle arrest mechanism by deletion of FAR1. This year, we adopt Fusion PCR method to construct linear recombinant fragment, as description of Figure 3.

Figure 3. A schematic diagram of the use of fusion PCR to synthesize a fragment for gene knockout. Flanking DNA fragments are amplified with primers P1 and P2 and with P5 and P6. Primers P3 and P4 are designed to amplify zeocin. The two flanking fragments and the zeocin fragment are mixed and fused together by PCR creating a linear fragment used for transformation. Finally, transforming the linear fragment into S.cerevisiae and homologous recombination will occur and delete the target gene.
Results
We delete the FAR1 gene successfully as shown in the figure 4.

Figure 4: An ethidium-biomide-stained agarose gel showing results of FAR1 gene knockout.
a: The first lane is a 250bp DNA marker ladder. The lanes 2-5 are products of PCR using p3 and p4 as the primers using the knockout Far1 yeast genome as the template but the lane 6 uses the original yeast genome as the template. The bands are correct size of zeocin which is not in the original S.cerevisiae BY4741.
b: The lane 6 is a 250bp DNA marker ladder. The lane 5 is product of PCR using primers inside Far1 using original yeast genome as the template but the remaining lanes use the knockout Far1 yeast genome as the template. The band is correct size of fragment inside Far1 gene.
Until now, we have successfully constructed 14 expression plasmids. The sequence of flag-tag, Rho and our olfactory receptor odr-10 has been optimized in genscript.

We have constructed three devices (DⅠ, DⅡ& DⅢ) to express the olfactory receptor(OR).
DⅠ: BBa_K1072013 
DⅡ: BBa_K1072014 
DⅢ: BBa_K1072015 
And then three odorant sensing systems(SⅠ, SⅡ& SⅢ) were inserted into shuttle plasmid YEplac181. Reporter device was BBa_K1072020, pFus1 + BFP + ADH1 terminator.
SⅠ:BBa_K1072016 
SⅡ: BBa_K1072017 
SⅢ: BBa_K1072018 
Our olfactory receptor odr-10 has been transformed into S.cerevisiae. we transformed SⅠBBa_K1072016 into S.cerevisiae and cultured the cells until they reached the exponential phase(OD600, in the range of 1-2), then we diluted broth to OD600 0.5 and cultured in the selection media containing 2% galactose at 30℃ for 18h.
Finally, we found that GFP, Flag tag signal and BFP were bright by confocal microscope,which means that Odr-10 gene have be translated, located and induced MAPK pathway successfully, as showing in figure 5.Flag tag was detected around plasma membrane, as we can see in figure 6.

Figure 5: Transforming Odr-10 with flag tag and reporter gene into yeast,GFP, flag tag signal and BFP was detected successfully, meaning that the whole pathway was stimulated by diacetyl.

Figure 6: FLAG immunolocalization of yeast cells expressing Odr-10 with flag tag, which means that Odr-10 protein locate at plasma membrane successfully.
Additionally, we transformed SⅡ system into S.cerevisiae and cultured the cells as above method. We found that SⅡsystem, BBa_K1072017, expresses bright blue fluorescence under UV field under 400ms exposure time Figure 7. Both BBa_K1072016 and BBa_K1072017 work meaning that Brho isn’t the necessary or don’t work in yeast. Odr-10 successful localization may result from the interior signal peptide of cDNA of Odr-10.

Figure 7.Transforming Odr-10 with flag tag and Brho into yeast, BFP brighten,meaning that the whole pathway work and demonstrate that Brho isn't the necessary factors expressing Odr-10 in yeast.
In order to optimize the pathway and achieve our expectation, we constructed Gpa1-odr3 chimera (chi) device to enhance the intension of signal transduction.
The chimera, carboxy-terminal five amino acids of Gpa1 are replaced with corresponding residues from odr-3 subunit, will not only enhance the affinity between Gpa1 with odr-10 from caenorhabditis elegans, but also not influence the affinity between Gpa1 with Gβγ. We utilize the sequence (1000bp) upstream of Gpa1 to serve as the pGpa1 and create a novel biobrick, BBa_K1072011. To make it can be adapted to principle of IGEM, we successfully mutate the restriction sites of EcoR1 and Spe1in the sequence through site-specific mutagenesis.
The chi device: 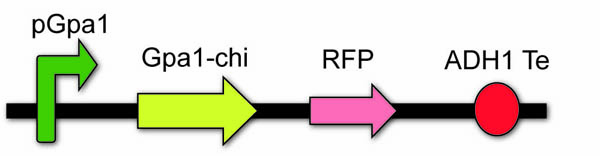
Reference
[1] Minic J, Persuy MA, Godel E et al. Functional expression of olfactory receptors in yeast and development of a bioassay for odorant screening. FEBS J. 2005 Jan;272(2):524-37.
[2] S. J. Dowell, A. J. Brown. Yeast Assays for G-Protein–Coupled Receptors. Receptors and Channels. 2002, Vol. 8, No. 5-6 , Pages 343-352.
[3] Audet M, Bouvier M. Restructuring G-protein-coupled receptor activation. Cell. 2012 Sep 28;151(1):14-23.
[4] Helen Dacres, Jian Wang,Virginia Leitch et al. Greatly enhanced detection of a volatile ligand at femtomolar levels using bioluminescence resonance energy transfer (BRET). Biosens Bioelectron. 2011 Nov 15;29(1):119-24.
[5] Radhika V, Proikas-Cezanne T, Jayaraman M et al. Chemical sensing of DNT by engineered olfactory yeast strain. Nat Chem Biol. 2007 Jun;3(6):325-30.
[6] Szewczyk E, Nayak T, Oakley CE, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1(6):3111-20.
[7] M. S. Ferry, I. A. Razinkov, and J. Hasty. Microfluidics for Synthetic Biology:From Design to Execution. Book, 2011.
 "
"



