Team:Carnegie Mellon/Project/Procedure
From 2013.igem.org
| (8 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
</ol> | </ol> | ||
<br><h3>Phage Construct</h3><p> | <br><h3>Phage Construct</h3><p> | ||
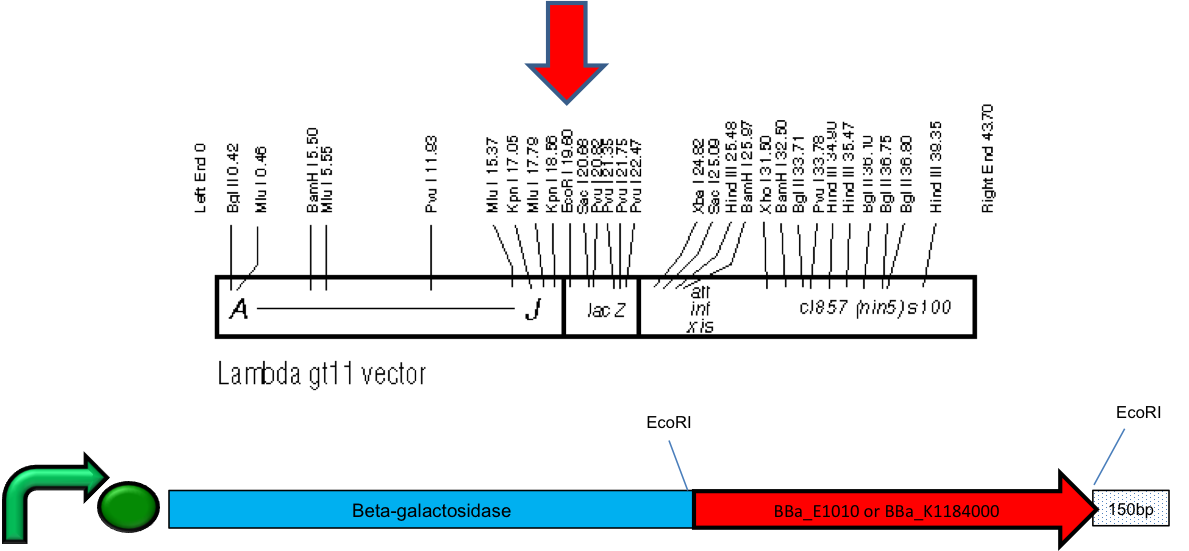
| - | The first goal was achieved using a lambda gt11 cloning kit and inserting the KillerRed and mRFP1 coding sequences into a β-galactosidase sequence, which results in a β-gal fusion protein to help with solubility and allow for blue, white screening. This also includes a single wild-type <i>lac</i> promoter, which responds to IPTG induction. Once clones were identified and isolated, the phages were then amplified using a strain of bacteria permissive for the lytic cycle. The mRFP1 fluorescence signal was detectable in plaques but KillerRed's fluorescence was barely above the autofluorescence of the media. Lysogens were made in the high-frequency-of-lysogeny (hfl) strain. Prophage are integrated into the genome and therefore single copy. KillerRed and mRFP1 lysogens showed no detectable fluorescence signal. This was repeated several times. The lysogen was then induced by shifting to | + | The first goal was achieved using a lambda gt11 cloning kit and inserting the KillerRed and mRFP1 coding sequences into a β-galactosidase sequence, which results in a β-gal fusion protein to help with solubility and allow for blue, white screening. This also includes a single wild-type <i>lac</i> promoter, which responds to IPTG induction. Once clones were identified and isolated, the phages were then amplified using a strain of bacteria permissive for the lytic cycle. The mRFP1 fluorescence signal was detectable in plaques but KillerRed's fluorescence was barely above the autofluorescence of the media. Lysogens were made in the high-frequency-of-lysogeny (hfl) strain. Prophage are integrated into the genome and therefore single copy. KillerRed and mRFP1 lysogens showed no detectable fluorescence signal. This was repeated several times. The lysogen was then induced by shifting to 42ºC to increase the copy number and very, very small amounts of mRFP were detected but no KillerRed. There was not enough protein for us to see killing by KillerRed. Ultimately, we were able to develop a lysogen that contains the gene for KillerRed but were having difficulty with expressing detectable amounts of protein as measured by fluorescence. </p></html> |
| + | [[image:Prophage.png|thumb|600px|center|Figure 1: The λ gt11 genome is shown above. The β-galactosidase gene is shown below with the EcoRI sites. The KillerRed and mRFP1 coding sequences are cloned into the EcoRI site.]] | ||
| + | <html> | ||
| + | <br><h3>Plasmid Construct</h3><p> | ||
<p> | <p> | ||
| - | In parallel, a plasmid construct (using pSB1A2) was made for easy, high level expression and to prove that KillerRed is able to kill bacteria. The plasmid construct consisted of a wild type <i>lac</i> promoter, like the phage and the 100% efficiency RBS standard as shown in Figure 1. Both KillerRed and mRFP1 were cloned so that the mRFP1 acted as the negative control. Induction with IPTG produced cultures that showed detectable expression of KillerRed and mRFP1, however the expression levels were consistently lower for KillerRed despite identical conditions and similar spectral properties. The maturation rate of KillerRed is longer than mRFP1 so it was kept at 4ºC overnight before photobleaching experiments. After sequence analysis, it was determined that the KillerRed is not codon optimized for <i>E. coli</i> and contains 13 rare proline codons, which may cause low translational efficiency and ultimately, low yield of protein expression. Despite this, photobleaching of both proteins were performed <i>in vivo</i> and the viable counts determined. Originally using various controls, it was determined that the lamp's light is non-toxic to the cells and therefore, killing can be attributed to KillerRed expression. As shown in our <a href="2013.igem.org/Team:Carnegie_Mellon/Project/Results">Results</a> page, we see a 50% decrease in cell viability after 5 hours of photobleaching. This demonstrated that KillerRed is capable of killing bacteria while the negative control, mRFP1, often grows in the presence of light. All of our photobleaching experiments were performed while the cells were log-phase growth, which may differ from previous literature protocols that report large percentages of killing. It is | + | In parallel, a plasmid construct (using pSB1A2) was made for easy, high level expression and to prove that KillerRed is able to kill bacteria. The plasmid construct consisted of a wild type <i>lac</i> promoter, like the phage and the 100% efficiency RBS standard as shown in Figure 1. Both KillerRed and mRFP1 were cloned so that the mRFP1 acted as the negative control. Induction with IPTG produced cultures that showed detectable expression of KillerRed and mRFP1, however the expression levels were consistently lower for KillerRed despite identical conditions and similar spectral properties. The maturation rate of KillerRed is longer than mRFP1 so it was kept at 4ºC overnight before photobleaching experiments. After sequence analysis, it was determined that the KillerRed is not codon optimized for <i>E. coli</i> and contains 13 rare proline codons, which may cause low translational efficiency and ultimately, low yield of protein expression. Despite this, photobleaching of both proteins were performed <i>in vivo</i> and the viable counts determined. Originally using various controls, it was determined that the lamp's light is non-toxic to the cells and therefore, killing can be attributed to KillerRed expression. As shown in our <a href="https://2013.igem.org/Team:Carnegie_Mellon/Project/Results">Results</a> page, we see a 50% decrease in cell viability after 5 hours of photobleaching. This demonstrated that KillerRed is capable of killing bacteria while the negative control for reactive oxygen production, mRFP1, often grows in the presence of light. All of our photobleaching experiments were performed while the cells were in log-phase growth, which may differ from previous literature protocols that report large percentages of killing. It is possible that superoxide dismutase is being synthesized in log-phase growth and therefore is able to limit the effect of the ROS. |
</p> | </p> | ||
</html> | </html> | ||
| - | [[image:Plasmid Construct.png|thumb|600px|center|Figure | + | [[image:Plasmid Construct.png|thumb|600px|center|Figure 2:The plasmid construct we use for expression consists of the <partinfo>BBa_R0010</partinfo> lac promoter, the <partinfo>BBa_B0034</partinfo> 100% efficiency RBS standard and either the KillerRed BBa_K1184000 coding sequence or mRFP1 <partinfo>BBa_E1010</partinfo> coding sequence. ]] |
<html> | <html> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | This is meant to be a guide to our experimental process, including challenges we encountered from conception to results. For a detailed description of our protocols and methods we used, visit our <a href="https://2013.igem.org/Team:Carnegie_Mellon/Protocols">Protocols</a> and <a href="https://2013.igem.org/Team:Carnegie_Mellon/Notebook">Notebook</a> pages. | + | <p>This is meant to be a guide to our experimental process, including challenges we encountered from conception to results. For a detailed description of our protocols and methods we used, visit our <a href="https://2013.igem.org/Team:Carnegie_Mellon/Protocols">Protocols</a> and <a href="https://2013.igem.org/Team:Carnegie_Mellon/Notebook">Notebook</a> pages. |
</html> | </html> | ||
| + | <br> | ||
| + | |||
| + | <h2>References</h2> | ||
| + | <p><sup>1</sup>Takemoto K, Matsuda T, Sakai N, Fu D, Noda M, Uchiyama S, Kotera I, Arai Y, Horiuchi M, Fukui K, Ayabe T, Inagaki F, Suzuki H, Nagai T. 2013. SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci Rep. 17(3):2629.</p> | ||
Latest revision as of 02:14, 28 September 2013
Procedure
There were two basic goals for this project:
- Engineer a temperate phage (specifically λ) to synthesize KillerRed in E. coli
- Demonstrate KillerRed's ability to kill E. coli
Phage Construct
The first goal was achieved using a lambda gt11 cloning kit and inserting the KillerRed and mRFP1 coding sequences into a β-galactosidase sequence, which results in a β-gal fusion protein to help with solubility and allow for blue, white screening. This also includes a single wild-type lac promoter, which responds to IPTG induction. Once clones were identified and isolated, the phages were then amplified using a strain of bacteria permissive for the lytic cycle. The mRFP1 fluorescence signal was detectable in plaques but KillerRed's fluorescence was barely above the autofluorescence of the media. Lysogens were made in the high-frequency-of-lysogeny (hfl) strain. Prophage are integrated into the genome and therefore single copy. KillerRed and mRFP1 lysogens showed no detectable fluorescence signal. This was repeated several times. The lysogen was then induced by shifting to 42ºC to increase the copy number and very, very small amounts of mRFP were detected but no KillerRed. There was not enough protein for us to see killing by KillerRed. Ultimately, we were able to develop a lysogen that contains the gene for KillerRed but were having difficulty with expressing detectable amounts of protein as measured by fluorescence.
Plasmid Construct
In parallel, a plasmid construct (using pSB1A2) was made for easy, high level expression and to prove that KillerRed is able to kill bacteria. The plasmid construct consisted of a wild type lac promoter, like the phage and the 100% efficiency RBS standard as shown in Figure 1. Both KillerRed and mRFP1 were cloned so that the mRFP1 acted as the negative control. Induction with IPTG produced cultures that showed detectable expression of KillerRed and mRFP1, however the expression levels were consistently lower for KillerRed despite identical conditions and similar spectral properties. The maturation rate of KillerRed is longer than mRFP1 so it was kept at 4ºC overnight before photobleaching experiments. After sequence analysis, it was determined that the KillerRed is not codon optimized for E. coli and contains 13 rare proline codons, which may cause low translational efficiency and ultimately, low yield of protein expression. Despite this, photobleaching of both proteins were performed in vivo and the viable counts determined. Originally using various controls, it was determined that the lamp's light is non-toxic to the cells and therefore, killing can be attributed to KillerRed expression. As shown in our Results page, we see a 50% decrease in cell viability after 5 hours of photobleaching. This demonstrated that KillerRed is capable of killing bacteria while the negative control for reactive oxygen production, mRFP1, often grows in the presence of light. All of our photobleaching experiments were performed while the cells were in log-phase growth, which may differ from previous literature protocols that report large percentages of killing. It is possible that superoxide dismutase is being synthesized in log-phase growth and therefore is able to limit the effect of the ROS.
This is meant to be a guide to our experimental process, including challenges we encountered from conception to results. For a detailed description of our protocols and methods we used, visit our Protocols and Notebook pages.
References
1Takemoto K, Matsuda T, Sakai N, Fu D, Noda M, Uchiyama S, Kotera I, Arai Y, Horiuchi M, Fukui K, Ayabe T, Inagaki F, Suzuki H, Nagai T. 2013. SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci Rep. 17(3):2629.
 "
"