Team:Yale/Project MAGE
From 2013.igem.org
(Difference between revisions)
(→Next steps) |
(→Next steps) |
||
| (7 intermediate revisions not shown) | |||
| Line 223: | Line 223: | ||
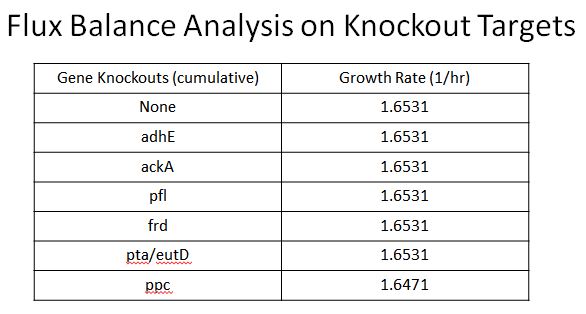
**These include adhE, ackA, pfl(A,B), frd(A,B,C,D, ppc, atoB, pta, eutD | **These include adhE, ackA, pfl(A,B), frd(A,B,C,D, ppc, atoB, pta, eutD | ||
**Oligos were designed to introduce two nonsence mutation near the begining of these ezymes, so they would not be expressed | **Oligos were designed to introduce two nonsence mutation near the begining of these ezymes, so they would not be expressed | ||
| - | **Using Flux Balance | + | **Using Flux Balance Analysis ([https://2013.igem.org/Team:Yale/Modeling instructions here]) we were able to determine that none of these enzymes would cause a fitness hit except ppc |
|style="padding-left: 20px; padding-right: 20px;"|[[File:FBAKO.JPG|400px]] | |style="padding-left: 20px; padding-right: 20px;"|[[File:FBAKO.JPG|400px]] | ||
|} | |} | ||
| Line 233: | Line 233: | ||
|- | |- | ||
| | | | ||
| + | |||
==== RBS Tuning ==== | ==== RBS Tuning ==== | ||
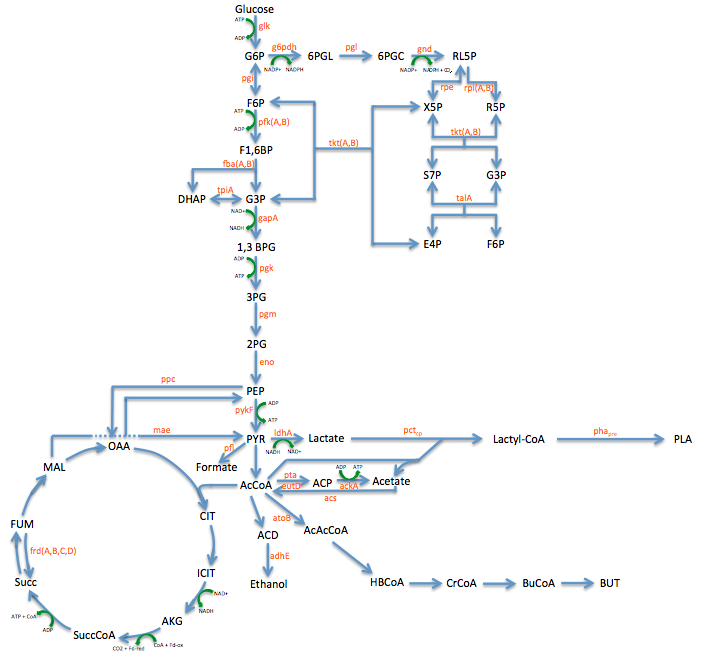
*Using the pathway diagram shown above we found 20 enzymes that could be potential targets for mutation | *Using the pathway diagram shown above we found 20 enzymes that could be potential targets for mutation | ||
| Line 246: | Line 247: | ||
== Results == | == Results == | ||
| - | *The first time we used FACS to sort the cells, we saw roughly a two fold increase in fluorescence when we tested on the plate reader | + | *The first time we used FACS to sort the cells, we saw roughly a two fold increase in fluorescence when we tested on the plate reader after only 1 round of MAGE with the KO oligos |
| - | + | ||
[[File:FACSsortingBandA.jpg|400px]] | [[File:FACSsortingBandA.jpg|400px]] | ||
| - | *We used this FACS sorted strain and ran 5 more MAGE cycles (one with RBS oligos, one with KO, and one with all Oligos) | + | *We used this 3 cultures of this FACS sorted strain and ran 5 more MAGE cycles each culture (one culture was MAGEd with the RBS oligos, one with KO, and one with all Oligos) |
**Here are the results of the FACS sorting of these strains | **Here are the results of the FACS sorting of these strains | ||
{| | {| | ||
|- | |- | ||
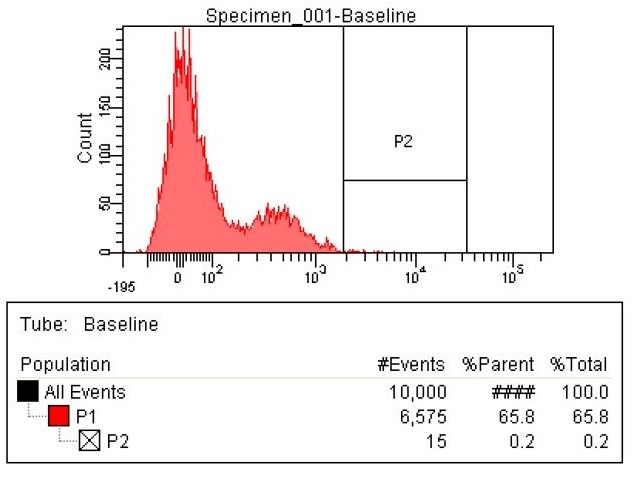
| - | |The first sample is the baseline. This is EcNR2 with our plasmid containing both the PCT and PHA gene. | + | |The first sample is the baseline. This is EcNR2 with our plasmid containing both the PCT and PHA gene. This strain was grown overnight with the cells induced and in the presence of Nile red to strain the PLA (this is the same procedure for all later strains as well). The gate (labeled P2) was chosen to select those with the highest levels of fluorescence. Due to the abnormally high levels of fluorescence in some cells in the baseline, the gate was chosen to include some baseline cells, but only those with very high levels of fluorescence. |
|style="padding-left: 20px; padding-right: 20px;"|[[File:Baseline both.jpg|400px]] | |style="padding-left: 20px; padding-right: 20px;"|[[File:Baseline both.jpg|400px]] | ||
|} | |} | ||
| Line 262: | Line 262: | ||
{| | {| | ||
|- | |- | ||
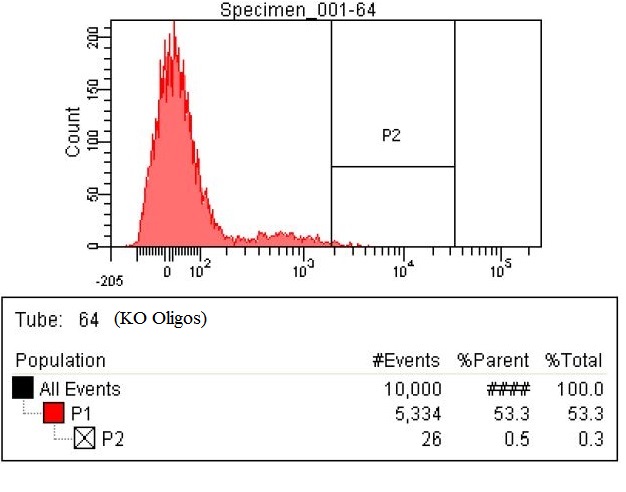
| - | |The second sample is the KO oligos. This means it | + | |The second sample is the KO oligos. This means it had a pool of 11 oligos to knock out 11 enzymes. The enzymes targeted for Knockout were ackA, frdB, frdD pflA, pflB, adhE, frdA, frdC, ATOB, PTA, EUTD. The chart shows that 26 cells within the P2 gate compared to the 15 in the baseline. That means number of cells with high levels of fluorescence nearly doubled. |
|style="padding-left: 20px; padding-right: 20px;"|[[File:64-KO knockouts both.jpg|400px]] | |style="padding-left: 20px; padding-right: 20px;"|[[File:64-KO knockouts both.jpg|400px]] | ||
|} | |} | ||
| Line 270: | Line 270: | ||
{| | {| | ||
|- | |- | ||
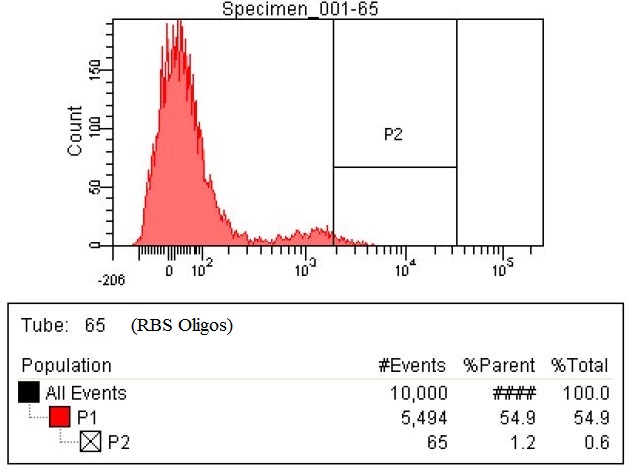
| - | |The third sample is the RBS tuning | + | |The third sample is the RBS tuning olgios. These are oligos have degenerate RBS site with the following sequence DDRRRRRDDDD (-4 through -14 positions from the start codon). The enzymes targeted for RBS tuning are DHA, ACS, ATOB, EUTD, PTA, MAEA, MAEB, PYKF, ENO, PGM, PGK, GAPA, TPIA, FBAA, FBAB, PFKA, PFKB, GLK, PGL, RPOS. The number of cells within the P2 gate is 65, which is greater than a fourfold increase compared to the baseline. |
|style="padding-left: 20px; padding-right: 20px;"|[[File:64-RBS both.jpg|400px]] | |style="padding-left: 20px; padding-right: 20px;"|[[File:64-RBS both.jpg|400px]] | ||
|} | |} | ||
| Line 278: | Line 278: | ||
{| | {| | ||
|- | |- | ||
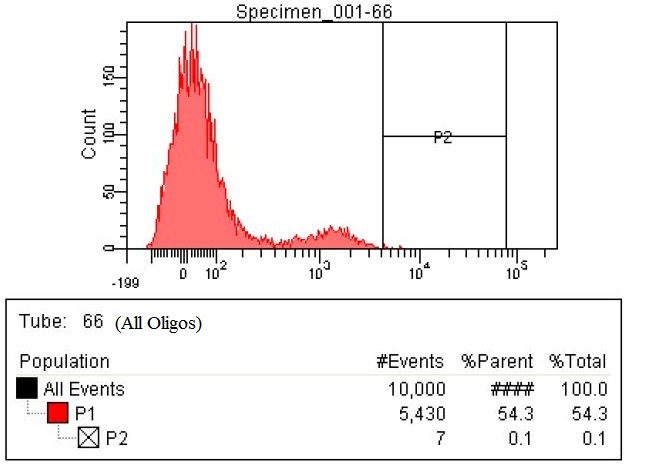
| - | |The fourth sample is the all | + | |The fourth sample is the all oligs. This is a combination of both the RBS oligos and the KO oligos. This sample has 98 cells within the gate, which is a six and a half fold increase. |
|style="padding-left: 20px; padding-right: 20px;"|[[File:66-All both.jpg|400px]] | |style="padding-left: 20px; padding-right: 20px;"|[[File:66-All both.jpg|400px]] | ||
|} | |} | ||
| Line 291: | Line 291: | ||
=== Next steps === | === Next steps === | ||
| - | *The next step is to grow up these cultures (we sorted | + | *The next step is to grow up these cultures (we sorted out 1,000 cells from each culture to start a culture). |
| - | *These cells will be tested on the plate reader compared to the wild type EcNR2, as well as the EcNR2 with our plasmid. | + | *These cells will be tested on the plate reader compared to the wild type EcNR2, as well as the wild type EcNR2 with our plasmid. The MAGEd and FACS sorted cells should show an increased levels of fluorescence, due to increased PLA production. With only 1 MAGE cycles we were able generate a twofold increase, so in theory with an additional 5 MAGE cycles this increase should be even more drastic. |
| - | + | ||
<br> | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
| - | + | ||
| + | == List of Papers == | ||
Jacob et al. 1997 <br> | Jacob et al. 1997 <br> | ||
Matsuzaki et al. 1998<br> | Matsuzaki et al. 1998<br> | ||
Latest revision as of 19:51, 8 October 2013
Contents |
MAGE Targets
- The first step in applying MAGE is finding MAGE targets. This involved reading numerous scientific papers to learn as much as possible about the heterologous enzymes, and the pathway that was being used to create the PLA
Enzyme Targets
- Sadly there was no crystal structure of either enzyme we could use to locate the sites to introduce mutations
- However, we used the literature available to locate spots where we would want to introduce mutations
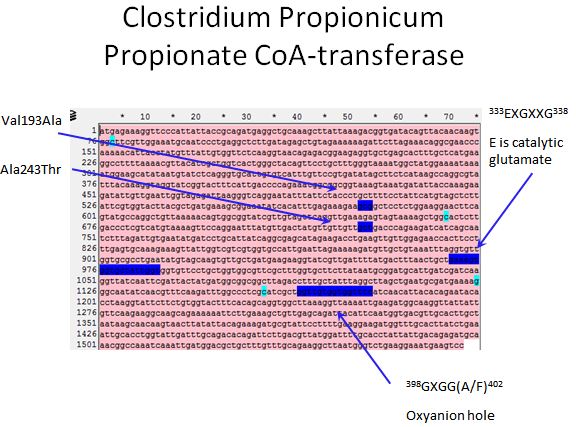
Propionate CoA-transferase
| 
|
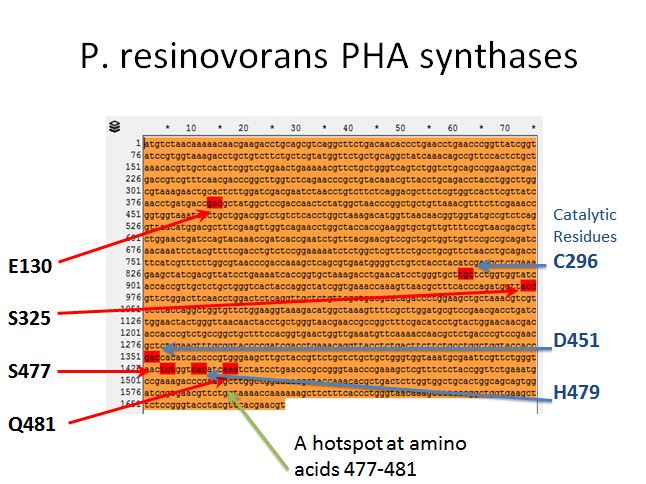
P. resinovorans PHA synthases
| 
|
Pathway Engineering
- We wanted to divert resources toward our desired pathway
- This mainly consisted of increasing the production of lactate
- In order to better understand the pathway we were tampering with we created this metabolic engineering graphic (using the sources listed at the bottom of this page)
Enzyme KOs
| 
|
RBS Tuning
| 
|
Results
- The first time we used FACS to sort the cells, we saw roughly a two fold increase in fluorescence when we tested on the plate reader after only 1 round of MAGE with the KO oligos
- We used this 3 cultures of this FACS sorted strain and ran 5 more MAGE cycles each culture (one culture was MAGEd with the RBS oligos, one with KO, and one with all Oligos)
- Here are the results of the FACS sorting of these strains
| The fourth sample is the all oligs. This is a combination of both the RBS oligos and the KO oligos. This sample has 98 cells within the gate, which is a six and a half fold increase. | 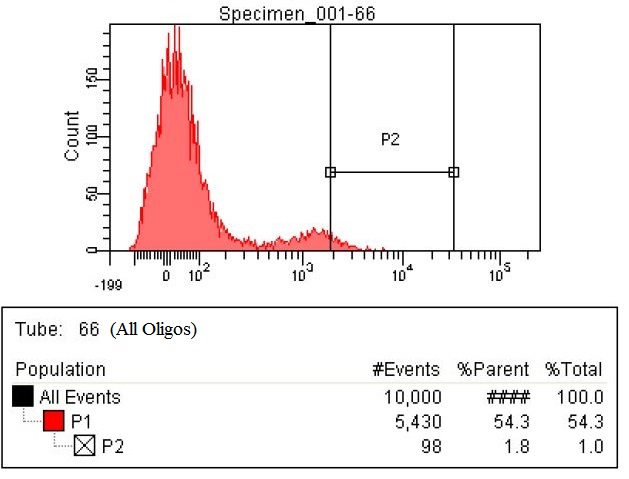
|
Next steps
- The next step is to grow up these cultures (we sorted out 1,000 cells from each culture to start a culture).
- These cells will be tested on the plate reader compared to the wild type EcNR2, as well as the wild type EcNR2 with our plasmid. The MAGEd and FACS sorted cells should show an increased levels of fluorescence, due to increased PLA production. With only 1 MAGE cycles we were able generate a twofold increase, so in theory with an additional 5 MAGE cycles this increase should be even more drastic.
List of Papers
Jacob et al. 1997
Matsuzaki et al. 1998
Sawers et al. 1998
Park et al. 2002
Selmer et al. 2002
Takase et al. 2002
Fong et al. 2005
Matsumoto et al. 2005
Rangarajan ES et al. 2005
Matsumoto et al. 2006
Jung et al. 2009
Matsumoto et al. 2009
Juang et al. 2010
Orth et al. 2010
Yang et al. 2011
Kandasamy et al. 2012
Yang et al. 2013
 "
"