Team:uOttawa/project
From 2013.igem.org
| (22 intermediate revisions not shown) | |||
| Line 13: | Line 13: | ||
<div class="row text-center pane intro"> | <div class="row text-center pane intro"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/ | + | <img src="https://static.igem.org/mediawiki/2013/7/7e/Uo2013-Wet_Lab_Info_v4.jpg" alt=""> |
</div> | </div> | ||
| Line 25: | Line 25: | ||
<img src="https://static.igem.org/mediawiki/2013/d/df/Uo2013-fflsimple.png" alt="Type-1 incoherent feed-forward loop" /> | <img src="https://static.igem.org/mediawiki/2013/d/df/Uo2013-fflsimple.png" alt="Type-1 incoherent feed-forward loop" /> | ||
</div> | </div> | ||
| - | <p>For this year’s project, we are aiming to construct an I1-FFL that can detect fold-changes in the levels of protein X. In other words, the expression of Z would be reliant on the relative change in X as opposed to absolute values of X. For example, if X changes from an arbitrary concentration of 1 to an arbitrary concentration of 5, the | + | <p>For this year’s project, we are aiming to construct an I1-FFL that can detect fold-changes in the levels of protein X. In other words, the expression of Z would be reliant on the relative change in X as opposed to absolute values of X. For example, if X changes from an arbitrary concentration of 1 to an arbitrary concentration of 5, the expression from promoter Z would be exactly the same as that resulting from a change from a concentration of 5 to a concentration of 25 of X (a fold-change by a factor of 5 in both cases).</p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 68: | Line 68: | ||
</div> | </div> | ||
| - | <div class=" | + | |
| - | <h1> | + | |
| - | <p> | + | |
| - | + | ||
| + | <div class="pane data"> | ||
| + | <div class="zebra"><div class="row"> | ||
| + | <h1>We have shown....</h1> | ||
| + | <ul class="bulleted"> | ||
| + | <li>Linearization of X activity is crucial to the function of the feedforward loop</li> | ||
| + | <li>Negative autoregulation can successfully linearize the dose-response of a downstream promoter</li> | ||
| + | <li>Transcriptional activators can be used as repressors and also act in negative auroregulation</li> | ||
| + | </ul> | ||
| + | </div></div> | ||
| + | |||
| + | <div class="zebra"><div class="row"> | ||
| + | <h1>Testing our basic network</h1> | ||
| + | <p>In the 2009 Alon paper, fold-change response to a stimulus was theoretically proposed. We set out to test this, by constructing a synthetic incoherent feedforward loop, as follows:</p> | ||
| + | <p class="text-center"><img src="https://static.igem.org/mediawiki/2013/6/64/UOttawa_project_image3.png" width="600px"/></p> | ||
| + | <p>Upon testing this network in a gradient of IPTG and ATc, we discovered that no evident pulse in sfGFP (Z) expression was observable in any range of the molecular cofactors upon stimulus of production of X (rtTA). This was explained by the Alon paper – in conditions of independent binding, as such in the case of our <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1164004" target="_blank">pTRELX promoter</a>, fold-change detection is only possible if the activity of the X activator on the Z promoter is linear.</p> | ||
| + | <p class="text-center" style="font-size: 14px">Click on the graph to expand</p> | ||
| + | <p class="text-center"><a href="https://static.igem.org/mediawiki/2013/e/e7/Uo2013-Gal1sfGFP_Graphs2.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2013/e/e7/Uo2013-Gal1sfGFP_Graphs2.jpg" /></a></p> | ||
| + | <p>The way the network is designed, the activity of the X activator on the Z promoter is sigmoidal, as is the case of most situations of activation. To combat this problem, we needed to find a way to linearize the activity of X on the Z promoter.</p> | ||
| + | </div></div> | ||
| + | |||
| + | <div class="zebra"><div class="row"> | ||
| + | <h1>How can we linearize the X activity?</h1> | ||
| + | <p>A 2009 paper by Balázsi illustrated how negative autoregulation can linearize the dose-response curve of a downstream promoter. We decided to attempt to incorporate negative autoregulation into our network by changing the promoter driving X (rtTA) production. By changing the promoter to pGalLX, which contains a lac operator site downstream of the TATA box. The downstream Y (LacI) is able to feedback to negatively regulate the production of rtTA. Since rtTA is what leads to production of LacI, any spikes in rtTA acvitity – as would be the case in a sigmoidal response – are dampened by increased presence of LacI. </p> | ||
| + | <p class="text-center"><img src="https://static.igem.org/mediawiki/2013/6/64/Uo2013-lxnetwork.jpg" width="600px" /></p> | ||
| + | <p>Upon testing these strains in a similar IPTG and ATc gradient, we were able to show a range of cofactor concentrations that allow for a temporal spike in Z (sfGFP) production, as expected from a fold-change response. The results of this experiment are shown below:</p> | ||
| + | <p class="text-center" style="font-size: 14px">Click on the graph to expand</p> | ||
| + | <p class="text-center"><a href="https://static.igem.org/mediawiki/2013/7/78/Uo2013-LXsfGFP_Graphs2.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2013/7/78/Uo2013-LXsfGFP_Graphs2.jpg" /></a></p> | ||
| + | </div></div> | ||
| + | |||
| + | <div class="zebra"><div class="row"> | ||
| + | <h1>Can we also use an activator to negatively regulate and linearize X activity?</h1> | ||
| + | <p>Although we were able to show that the downstream LacI is able to negatively regulate and linearize rtTA activity, we wanted to determine whether this would be possible with rtTA itself. Such a situation would theoretically allow for tighter regulation of rtTA activity, since no downstream transcription factors would be involved. To test this, we replaced the pGal1 promoter in the original network with pGalTX, which contains two tet operators downstream of the TATA box. Although rtTA is an activator, binding of the activator downstream of the TATA box should theoretically prevent transcription via steric hindrance. Again, spikes in rtTA activity should be dampened by binding of rtTA itself to its own promoter.</p> | ||
| + | <p class="text-center"><img src="https://static.igem.org/mediawiki/2013/f/f8/Uo2013-txnetwork.jpg" width="600px"/></p> | ||
| + | <p>When we tested these strains in the IPTG and ATc gradient, we observed a very small range of cofactor concentrations that allowed for the temporal spike in sfGFP.</p> | ||
| + | <p class="text-center" style="font-size: 14px">Click on the graph to expand</p> | ||
| + | <p class="text-center"><a href="https://static.igem.org/mediawiki/2013/6/6d/Uo2013-TXsfGFP_Graphs2.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2013/6/6d/Uo2013-TXsfGFP_Graphs2.jpg" /></a></p> | ||
| + | <p>With tighter regulation of the rtTA activity, we would expect stronger negative regulation and thus tighter linearization of rtTA activity, however, this is not what is observed in our results. Here, we observe a more narrow window of cofactor concentrations that allow for the temporal pulse in sfGFP. This may have to do with the sequestering of the rtTA protein to its own promoter instead of the downstream promoters. As the rtTA is now effectively regulating the activity of all three promoters that are crucial to the function of the feedforward loop infrastructure, it makes sense that the regulation of its binding affinity must be more tightly controlled than in previous cases for successful function of the network.</p> | ||
| + | </div></div> | ||
| + | |||
| + | <div class="zebra"><div class="row"> | ||
| + | <h1>Future directions</h1> | ||
| + | <p>Now that we have determined that our incoherent feedforward loop is functional as expected and is able to produce a temporal Z (sfGFP) response, we will be able to test the fold-change capabilities of our synthetic network by regulating the concentration of the X (rtTA) stimulus over time.</p> | ||
| + | </div></div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
<div class="pane collabs"> | <div class="pane collabs"> | ||
| Line 83: | Line 131: | ||
</div></div> | </div></div> | ||
<div class="zebra"><div class="row"> <h1>Collaboration: Purdue</h1> | <div class="zebra"><div class="row"> <h1>Collaboration: Purdue</h1> | ||
| - | <p>In addition to our wet lab collaboration with Waterloo, we were also able to participate in the largest collaborative project in the history of iGEM, in partnership with Purdue University. Purdue contacted many iGEM teams across all corners of the world, and together we worked to propose a new characterization standard for the Registry of Standard Biological Parts. Thanks for the invitation, Purdue! <a href="https://2013.igem.org/Team:Purdue/Human_Practices/iGEM_Characterization_Standard">Learn more.</a></p> | + | <p>In addition to our wet lab collaboration with Waterloo, we were also able to participate in the largest collaborative project in the history of iGEM, in partnership with Purdue University. Purdue contacted many iGEM teams across all corners of the world, and together we worked to propose a new characterization standard for the Registry of Standard Biological Parts. Thanks for the invitation, Purdue! <a target="_blank" href="https://2013.igem.org/Team:Purdue/Human_Practices/iGEM_Characterization_Standard">Learn more.</a></p> |
<img src="https://static.igem.org/mediawiki/2013/9/92/IWorld_Map.png" width="1100" height="200"> | <img src="https://static.igem.org/mediawiki/2013/9/92/IWorld_Map.png" width="1100" height="200"> | ||
</div></div> | </div></div> | ||
| Line 90: | Line 138: | ||
<div class="row pane references"> | <div class="row pane references"> | ||
<h1>References</h1> | <h1>References</h1> | ||
| - | < | + | <ul class="refList"><li>1. <a target="_blank" href="http://www.ncbi.nlm.nih.gov/pubmed/23676681">Daniel R, Rubens JR, Sarpeshkar R, Lu TK (2013) Synthetic analog computation in living cells. Nature 497: 619–623</a></li> |
| - | < | + | <li>2. <a target="_blank" href="http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2680460/">Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol. 2009;27:465</a></li> |
| - | < | + | <li>3. <a target="_blank" href="http://www.ncbi.nlm.nih.gov/pubmed/19363495">Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., 3rd., Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318. </a></li> |
| - | < | + | <li>4. <a target="_blank" href="http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2896310/">Goentoro L, Shoval O, Kirschner MW, Alon U (2009) The incoherent feedforward loop can provide fold-change detection in gene regulation. Molecular Cell 36: 894–899.</a></li> |
| - | < | + | <li>5. <a target="_blank" href="http://www.ncbi.nlm.nih.gov/pubmed/2744487">Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-Directed Mutagenesis by Overlap Extension Using the Polymerase Chain-Reaction. Gene 77: 51–59. </a></li> |
| - | < | + | <li>6. <a target="_blank" href="http://www.ruor.uottawa.ca/en/handle/10393/24000">Jedrysiak, Daniel K. "Construction and Characterization of Gene Regulatory Networks in Yeast." Order No. MR86054 University of Ottawa (Canada), 2013. Ann Arbor: ProQuest. Web. 27 Sep. 2013. </a></li> |
| - | < | + | <li>7. <a target="blank" href="http://www.nature.com/nbt/journal/v30/n7/full/nbt.2281.html?WT.ec_id=NBT-201207">Khmelinskii, A., P.J. Keller, A. Bartosik, M. Meurer, J.D. Barry, B.R. Mardin, A. Kaufmann, S. Trautmann, M. Wachsmuth, G. Pereira, W. Huber, E. Schiebel and M. Knop (2012). Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat. Biotechnol., 30:708-714.</a></li> |
| - | < | + | <li>8. <a target="_blank" href="http://www.ncbi.nlm.nih.gov/pubmed/20810924">Kuttykrishnan S, Sabina J, Langton LL, Johnston M, Brent MR. A quantitative model of glucose signaling in yeast reveals an incoherent feed forward loop leading to a specific, transient pulse of transcription. Proc Natl Acad Sci USA. 2010;107:16743–16748. doi: 10.1073/pnas.0912483107. </a></li> |
| - | < | + | <li>9. <a target="_blank" href="http://www.sysbiolab.uottawa.ca/publications/Kaern_SMCB_2006.pdf">Mads Kærn & Ron Weiss. Synthetic gene regulatory systems. In: Systems Modeling in Cellular Biology. Z. Sallasi et al. (Eds.). MIT Press (2006). </a></li> |
| - | < | + | <li>10. <a target="_blank" href="http://www.ncbi.nlm.nih.gov/pubmed/14530388">Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–85. Pioneering study of the functional significance of network motifs.</a></li> |
| - | < | + | <li>11. <a target="_blank" href="http://www.ncbi.nlm.nih.gov/pubmed/21965290">McIsaac RS, et al. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Molecular biology of the cell. 2011;22:4447–4459. </a></li> |
| - | < | + | <li>12. <a target="_blank" href="http://www.ncbi.nlm.nih.gov/pubmed/19279212">Nevozhay D, Adams RM, Murphy KF, Josic K, Balázsi G. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci USA. 2009;106(13):5123–5128.</a></li></ul> |
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 03:49, 29 October 2013

Background
The Type-I Incoherent Feedforward Loop and Fold-Change Detection
The type-I incoherent feedforward loop (I1-FFL) is a gene network in which protein X activates a gene Z while simultaneously activating the production of a repressor of gene Z, labeled Y.

For this year’s project, we are aiming to construct an I1-FFL that can detect fold-changes in the levels of protein X. In other words, the expression of Z would be reliant on the relative change in X as opposed to absolute values of X. For example, if X changes from an arbitrary concentration of 1 to an arbitrary concentration of 5, the expression from promoter Z would be exactly the same as that resulting from a change from a concentration of 5 to a concentration of 25 of X (a fold-change by a factor of 5 in both cases).
How Fold-Change Detection is Achieved
This fold-change detection is enabled through the repressor Y. Take the case when activator X, repressor Y, and protein Z each have an arbitrary concentration of 1 in the cell (1:1:1 ratio). If the concentration of X is doubled, the ratio between X and Y now becomes 2:1. X immediately activates Z, but since the repressor Y takes time to fold, it lags behind, and the concentration of Z spikes to 2 due to the activation. When Y folds into its active form and carries out its repressive activity, the concentration of Z returns to its initial concentration of 1. This is because at this point, X and Y return to a 1:1 ratio in the cell. In order for the concentration of Z to reach 2 once again, the X:Y ratio must first return to 2:1 - thus, X has to reach a concentration of 4 before the same output of Z is produced. In this way, fold-change detection is achieved.

Engineering our System
The Design of our Gene Network and the Interactions Between Components
In our proposed design, rtTA will act as our activator (X), Lacl as our repressor (Y), and sfGFP as our reporter protein (Z). Each part is detectable via a different fluorescent protein. A visual representation of our pathway is shown in Figure 3.
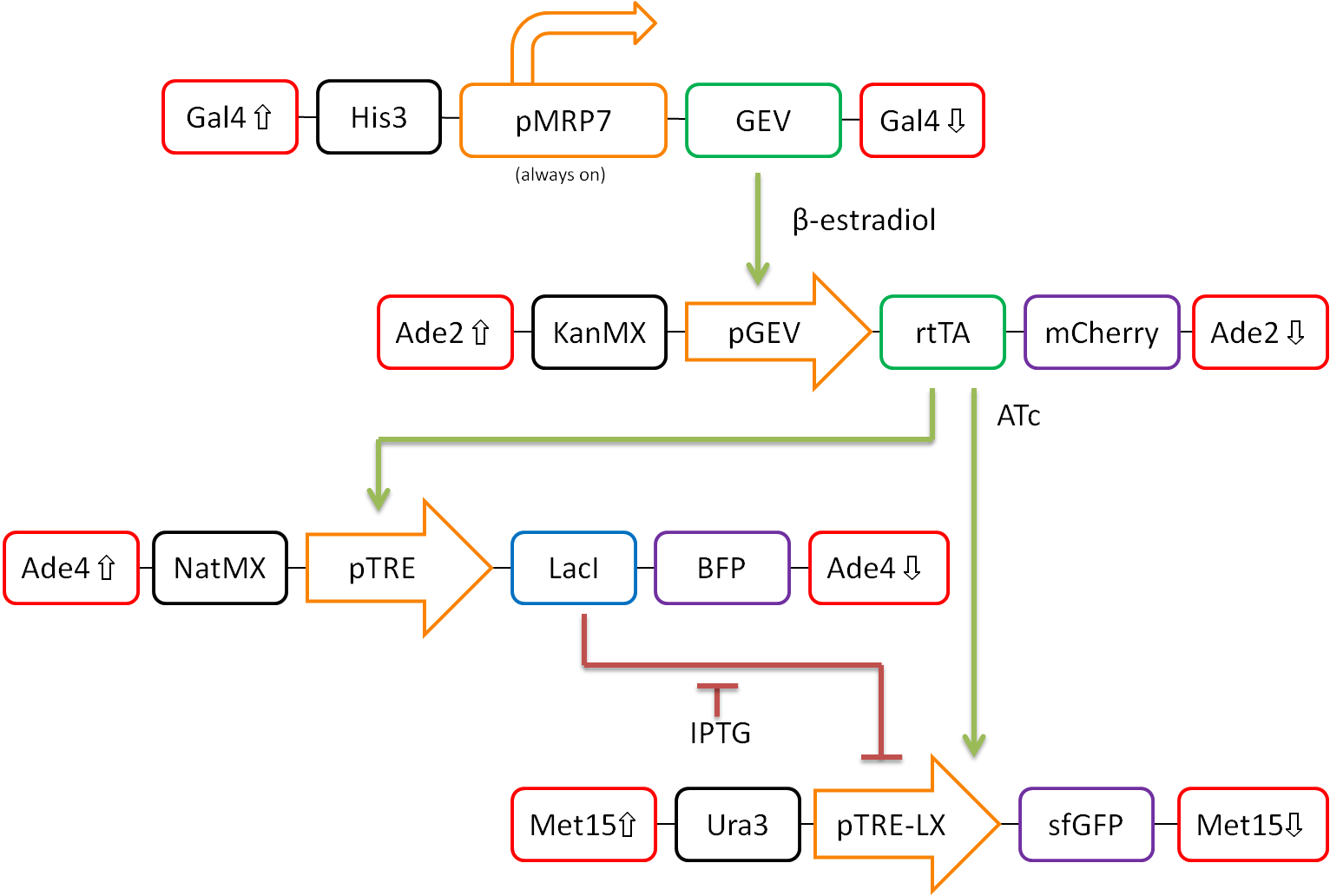
This network design allows for calibration of the network via tunable levels of β-estradiol, allowing for accurate control of the amount of rtTA(X) in the cell. This will allow us to ensure that fold-change detection works as theorized in our network.
Furthermore, activation by rtTA is mediated by varying levels of anhydrotetracycline (ATc), and LacI repression is mediated by varying levels of Isopropyl β-D-1-thiogalactopyranoside (IPTG). These methods of mediation allow for high tunability of the system, and we should be able to find a concentration range of ATc and IPTG at which the system will work as desired.
The system will then be characterized extensively. The fluorescent markers tagged to each component of the network will allow for simple temporal analysis using flow cytometry. This will allow for a high-throughput analysis of the activity of our network.
Potential Applications
A Fold-Change Detection System for Toxic Molecules
Once our system is tuned and is working, it can be modified to act as a fold-change detector for toxic molecules. By replacing the pGEV promoter in front of the rtTA gene with a promoter that is inducible by a toxic molecule, the amount of rtTA produced will be directly correlated to the concentration of the toxic molecule in the cellular environment.

This detector system is advantageous in that it detects fold-changes rather than changes in absolute values, which gives meaning to a signal in reference to the background signal level. This allows for the network to respond only to a signal that rises significantly above the background noise.
We have shown....
- Linearization of X activity is crucial to the function of the feedforward loop
- Negative autoregulation can successfully linearize the dose-response of a downstream promoter
- Transcriptional activators can be used as repressors and also act in negative auroregulation
Testing our basic network
In the 2009 Alon paper, fold-change response to a stimulus was theoretically proposed. We set out to test this, by constructing a synthetic incoherent feedforward loop, as follows:

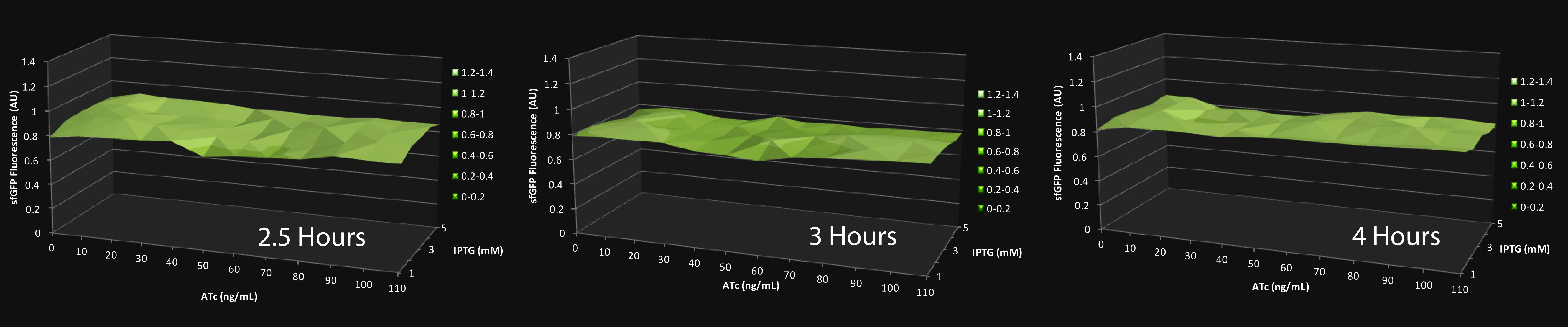
Upon testing this network in a gradient of IPTG and ATc, we discovered that no evident pulse in sfGFP (Z) expression was observable in any range of the molecular cofactors upon stimulus of production of X (rtTA). This was explained by the Alon paper – in conditions of independent binding, as such in the case of our pTRELX promoter, fold-change detection is only possible if the activity of the X activator on the Z promoter is linear.
Click on the graph to expand
The way the network is designed, the activity of the X activator on the Z promoter is sigmoidal, as is the case of most situations of activation. To combat this problem, we needed to find a way to linearize the activity of X on the Z promoter.
How can we linearize the X activity?
A 2009 paper by Balázsi illustrated how negative autoregulation can linearize the dose-response curve of a downstream promoter. We decided to attempt to incorporate negative autoregulation into our network by changing the promoter driving X (rtTA) production. By changing the promoter to pGalLX, which contains a lac operator site downstream of the TATA box. The downstream Y (LacI) is able to feedback to negatively regulate the production of rtTA. Since rtTA is what leads to production of LacI, any spikes in rtTA acvitity – as would be the case in a sigmoidal response – are dampened by increased presence of LacI.

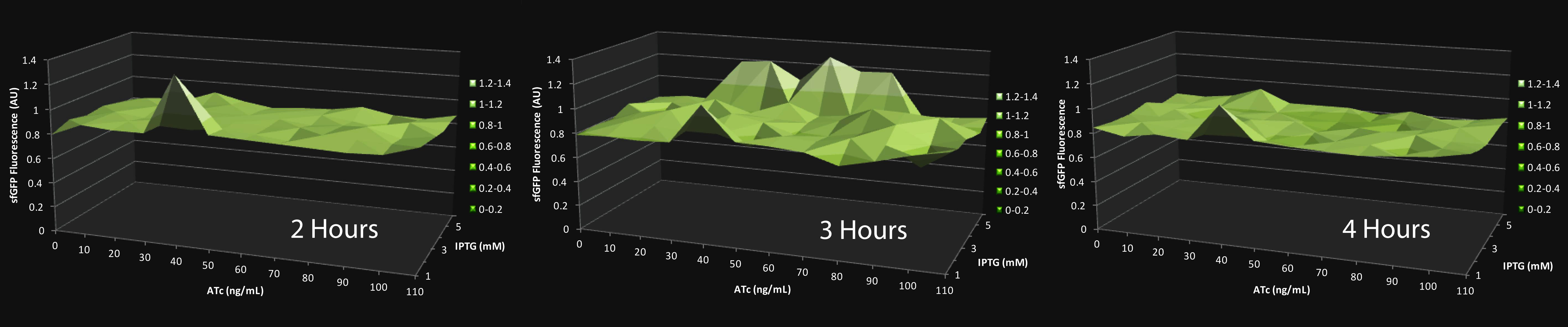
Upon testing these strains in a similar IPTG and ATc gradient, we were able to show a range of cofactor concentrations that allow for a temporal spike in Z (sfGFP) production, as expected from a fold-change response. The results of this experiment are shown below:
Click on the graph to expand
Can we also use an activator to negatively regulate and linearize X activity?
Although we were able to show that the downstream LacI is able to negatively regulate and linearize rtTA activity, we wanted to determine whether this would be possible with rtTA itself. Such a situation would theoretically allow for tighter regulation of rtTA activity, since no downstream transcription factors would be involved. To test this, we replaced the pGal1 promoter in the original network with pGalTX, which contains two tet operators downstream of the TATA box. Although rtTA is an activator, binding of the activator downstream of the TATA box should theoretically prevent transcription via steric hindrance. Again, spikes in rtTA activity should be dampened by binding of rtTA itself to its own promoter.

When we tested these strains in the IPTG and ATc gradient, we observed a very small range of cofactor concentrations that allowed for the temporal spike in sfGFP.
Click on the graph to expand
With tighter regulation of the rtTA activity, we would expect stronger negative regulation and thus tighter linearization of rtTA activity, however, this is not what is observed in our results. Here, we observe a more narrow window of cofactor concentrations that allow for the temporal pulse in sfGFP. This may have to do with the sequestering of the rtTA protein to its own promoter instead of the downstream promoters. As the rtTA is now effectively regulating the activity of all three promoters that are crucial to the function of the feedforward loop infrastructure, it makes sense that the regulation of its binding affinity must be more tightly controlled than in previous cases for successful function of the network.
Future directions
Now that we have determined that our incoherent feedforward loop is functional as expected and is able to produce a temporal Z (sfGFP) response, we will be able to test the fold-change capabilities of our synthetic network by regulating the concentration of the X (rtTA) stimulus over time.
Collaboration: Waterloo
This year, we had the opportunity to collaborate with the iGEM team from the University of Waterloo. We were asked to aid them in the construction of four large DNA constructs for their project, as illustrated below:

In order to do so, the uOttawa iGEM Team designed a total of 36 primers for the uWaterloo team. Because many of the individual parts were very short, an experimental oligonucleotide-based synthesis method was used, in combination with a modified overlap extension PCR assembly method. A very detailed protocol was written for the uWaterloo team, and they were successfully able to construct their DNA constructs using our method.
In exchange for the construction aid, the uWaterloo team compiled a list of differential equations that were used to model our synthetic biological system. The equations were also implemented in MATLAB for us by the uWaterloo team. Thanks for the incredible help, Paul and the uWaterloo team! Learn more.
Collaboration: Purdue
In addition to our wet lab collaboration with Waterloo, we were also able to participate in the largest collaborative project in the history of iGEM, in partnership with Purdue University. Purdue contacted many iGEM teams across all corners of the world, and together we worked to propose a new characterization standard for the Registry of Standard Biological Parts. Thanks for the invitation, Purdue! Learn more.

References
- 1. Daniel R, Rubens JR, Sarpeshkar R, Lu TK (2013) Synthetic analog computation in living cells. Nature 497: 619–623
- 2. Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol. 2009;27:465
- 3. Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., 3rd., Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318.
- 4. Goentoro L, Shoval O, Kirschner MW, Alon U (2009) The incoherent feedforward loop can provide fold-change detection in gene regulation. Molecular Cell 36: 894–899.
- 5. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-Directed Mutagenesis by Overlap Extension Using the Polymerase Chain-Reaction. Gene 77: 51–59.
- 6. Jedrysiak, Daniel K. "Construction and Characterization of Gene Regulatory Networks in Yeast." Order No. MR86054 University of Ottawa (Canada), 2013. Ann Arbor: ProQuest. Web. 27 Sep. 2013.
- 7. Khmelinskii, A., P.J. Keller, A. Bartosik, M. Meurer, J.D. Barry, B.R. Mardin, A. Kaufmann, S. Trautmann, M. Wachsmuth, G. Pereira, W. Huber, E. Schiebel and M. Knop (2012). Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat. Biotechnol., 30:708-714.
- 8. Kuttykrishnan S, Sabina J, Langton LL, Johnston M, Brent MR. A quantitative model of glucose signaling in yeast reveals an incoherent feed forward loop leading to a specific, transient pulse of transcription. Proc Natl Acad Sci USA. 2010;107:16743–16748. doi: 10.1073/pnas.0912483107.
- 9. Mads Kærn & Ron Weiss. Synthetic gene regulatory systems. In: Systems Modeling in Cellular Biology. Z. Sallasi et al. (Eds.). MIT Press (2006).
- 10. Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–85. Pioneering study of the functional significance of network motifs.
- 11. McIsaac RS, et al. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Molecular biology of the cell. 2011;22:4447–4459.
- 12. Nevozhay D, Adams RM, Murphy KF, Josic K, Balázsi G. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci USA. 2009;106(13):5123–5128.
 "
"