Team:Northwestern/methods
From 2013.igem.org
2013.igem.nu (Talk | contribs) |
2013.igem.nu (Talk | contribs) |
||
| (6 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
p { | p { | ||
| + | text-indent: 30px; | ||
color:black; | color:black; | ||
font-family: helvetica; | font-family: helvetica; | ||
} | } | ||
| + | a { | ||
| + | color:black; | ||
| + | font-family: helvetica; } | ||
.container { | .container { | ||
| Line 132: | Line 136: | ||
color: #6A1A7E; | color: #6A1A7E; | ||
font-family: fantasy; | font-family: fantasy; | ||
| + | border-bottom-width: thick; | ||
| + | border-bottom-style: solid; | ||
| + | border-bottom-color: #6A1A7E; | ||
| + | } | ||
| + | |||
| + | h2{ | ||
| + | color: #6A1A7E; | ||
| + | font-family: fantasy; | ||
| + | font-size: 20px; | ||
border-bottom-width: thick; | border-bottom-width: thick; | ||
border-bottom-style: solid; | border-bottom-style: solid; | ||
| Line 175: | Line 188: | ||
</style> | </style> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | </br> | ||
<div> | <div> | ||
<h1> Methods Overview </h1> | <h1> Methods Overview </h1> | ||
<div> | <div> | ||
| - | + | <center> <img src="https://static.igem.org/mediawiki/2013/6/6d/Dual-state.png"/> </center> | |
| + | |||
| + | <p>Our dual-state construct consists of a pH-inducible promoter, joined in series with a nonsense spacer region, a constitutive promoter, a ribosomal binding site, and a green-fluorescent protein capped with a terminator, as diagrammed above.</p> | ||
| + | |||
| + | <p>In creating our dual-state constructs, we worked with a low-copy plasmid, plasmid pSB4A5 taken from iGEM 2013’s distribution kit. This plasmid was taken from Kit Plate 5, well 21F. We wanted to ensure that our engineered cells would survive with our constructs, as they would need to survive should our construct be implemented as intended. We used a low-copy plasmid, because a high-copy plasmid would put too much stress on our cells and hinder their survival.</p> | ||
| + | |||
| + | <p>The table below summarizes the combinations of pH-inducible promoters, spacers, and constitutive promoters we attempted to construct into dual-state promoters.</p> | ||
| + | |||
| + | <center> <img src="https://static.igem.org/mediawiki/2013/4/4a/Dual_state_table.png"/> </center> | ||
| + | |||
| + | <center> <p><b>The diagrams below demonstrate our cloning strategy in creating these dual-state promoters.<b></p> </center> | ||
| + | |||
| + | <p><b>Stage 1: </b> We began by creating constructs containing Asr-rbs, GadA-rbs, Lpp-RBS, and TacRBS in conjunction with GFP and a terminator sequence in order to test the fluorescence of individual promoters at different pH levels (pH 3.5, 4.5, 5.5, 6.5, 7.5). This allows us to compare activities of these promoters in the dual-state against their activities in the single-state to better assess how these promoters might operate differently in a dual-state.</p> | ||
| + | |||
| + | <center> <img src="https://static.igem.org/mediawiki/2013/e/ef/Diagram1.png" height="400" width="500"/></center> | ||
| + | |||
| + | <p><b>Stage 2a: </b> We proceeded by placing each of our pH-inducible promoters upstream of the each of our nonsense spacer regions.</p> | ||
| + | <p> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2013/a/af/Diagram2.png" height="350" width="500" /></center> | ||
| + | |||
| + | Stage 2b: Finally, we were able to construct our dual-state promoters. We placed our pH-inducible promoter+spacer region constructs upstream constitutive promoter+RBS+GFP+terminator constructs. We were able to obtain these constitutive promoter+RBS+GFP+terminator constructs from work our TA Jessica Perez did separately as a member of the Jewett Laboratory at Northwestern University. We characterized these constructs by performing a fluorescence assay with minimal growth media buffered to pH 3.5, 4.5, 5.5, 6.5, and 7.5.</p> | ||
| + | |||
| + | <center><img src="https://static.igem.org/mediawiki/2013/e/e2/Diagram3.png" height="400" width="400" /> </center> | ||
| + | |||
| + | |||
</div> | </div> | ||
<h1>Strains and Media</h1> | <h1>Strains and Media</h1> | ||
| - | <p> | + | <p>Escherichia coli Top10 (Invitrogen) was used for all transformations and assays. Media included SOB for transformations and LB for overnight cultures. Transformed strains were grown at 37°C using Ampicillin resistance. Primers for PCR were purchased from Integrated DNA Technologies (IDT) and New England Biolabs (NEB) donated all of the restriction enzymes. All sequencing was conducted by the Northwestern Genomics Core.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<h1> Protocols </h1> | <h1> Protocols </h1> | ||
<div> | <div> | ||
| - | <h2> Supplies </h2> | + | <h2> Supplies </h2><center> |
<p> <a href="https://2013.igem.org/Team:Northwestern/makem9">Making M9 Media</a></p> | <p> <a href="https://2013.igem.org/Team:Northwestern/makem9">Making M9 Media</a></p> | ||
<p> <a href="https://2013.igem.org/Team:Northwestern/makecells">Making Competent Cells</a></p> | <p> <a href="https://2013.igem.org/Team:Northwestern/makecells">Making Competent Cells</a></p> | ||
<p> <a href="https://2013.igem.org/Team:Northwestern/transform">Transforming Competent Cells</a></p> | <p> <a href="https://2013.igem.org/Team:Northwestern/transform">Transforming Competent Cells</a></p> | ||
| - | <p> <a href="https://2013.igem.org/Team:Northwestern/lb">Making LB Plates</a></p> | + | <p> <a href="https://2013.igem.org/Team:Northwestern/lb">Making LB Plates</a></p></center> |
| - | <h2> | + | <h2> Assay </h2><center> |
| + | <p> <a href="https://2013.igem.org/Team:Northwestern/fluo">Fluorescence Assay</a></p></center> | ||
| + | <h2> Cloning </h2><center> | ||
<p> <a href="https://2013.igem.org/Team:Northwestern/primers">Annealing Primers</a></p> | <p> <a href="https://2013.igem.org/Team:Northwestern/primers">Annealing Primers</a></p> | ||
<p> <a href="https://2013.igem.org/Team:Northwestern/pcr">PCR Amplification</a></p> | <p> <a href="https://2013.igem.org/Team:Northwestern/pcr">PCR Amplification</a></p> | ||
| Line 230: | Line 241: | ||
<p> <a href="https://2013.igem.org/Team:Northwestern/digest">Restriction Digest</a></p> | <p> <a href="https://2013.igem.org/Team:Northwestern/digest">Restriction Digest</a></p> | ||
<p> <a href="https://2013.igem.org/Team:Northwestern/ligate">Ligation</a></p> | <p> <a href="https://2013.igem.org/Team:Northwestern/ligate">Ligation</a></p> | ||
| - | + | </center> | |
| - | + | ||
| - | + | ||
</div> | </div> | ||
Latest revision as of 04:04, 28 September 2013
Methods Overview

Our dual-state construct consists of a pH-inducible promoter, joined in series with a nonsense spacer region, a constitutive promoter, a ribosomal binding site, and a green-fluorescent protein capped with a terminator, as diagrammed above.
In creating our dual-state constructs, we worked with a low-copy plasmid, plasmid pSB4A5 taken from iGEM 2013’s distribution kit. This plasmid was taken from Kit Plate 5, well 21F. We wanted to ensure that our engineered cells would survive with our constructs, as they would need to survive should our construct be implemented as intended. We used a low-copy plasmid, because a high-copy plasmid would put too much stress on our cells and hinder their survival.
The table below summarizes the combinations of pH-inducible promoters, spacers, and constitutive promoters we attempted to construct into dual-state promoters.

The diagrams below demonstrate our cloning strategy in creating these dual-state promoters.
Stage 1: We began by creating constructs containing Asr-rbs, GadA-rbs, Lpp-RBS, and TacRBS in conjunction with GFP and a terminator sequence in order to test the fluorescence of individual promoters at different pH levels (pH 3.5, 4.5, 5.5, 6.5, 7.5). This allows us to compare activities of these promoters in the dual-state against their activities in the single-state to better assess how these promoters might operate differently in a dual-state.

Stage 2a: We proceeded by placing each of our pH-inducible promoters upstream of the each of our nonsense spacer regions.
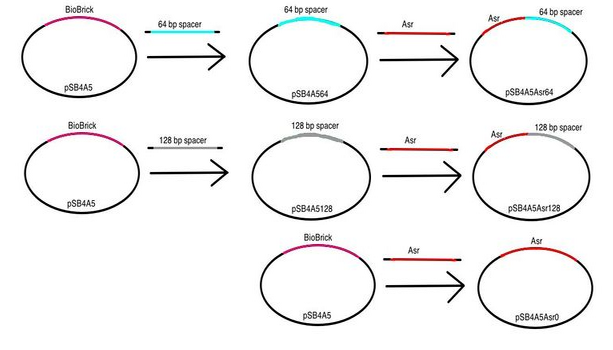

Strains and Media
Escherichia coli Top10 (Invitrogen) was used for all transformations and assays. Media included SOB for transformations and LB for overnight cultures. Transformed strains were grown at 37°C using Ampicillin resistance. Primers for PCR were purchased from Integrated DNA Technologies (IDT) and New England Biolabs (NEB) donated all of the restriction enzymes. All sequencing was conducted by the Northwestern Genomics Core.
 "
"
