Team:Greensboro-Austin/ncAAs
From 2013.igem.org
m |
(→Efficiency) |
||
| (12 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Greensboro-Austin/Stylesheet}} | {{Template:Greensboro-Austin/Stylesheet}} | ||
| - | + | __NOTOC__ | |
<html> | <html> | ||
| Line 17: | Line 17: | ||
<li class="parts_submitted"><a href="/Team:Greensboro-Austin/Parts" title="parts_submitted"><span class="displace">Parts Submitted</span></a></li> | <li class="parts_submitted"><a href="/Team:Greensboro-Austin/Parts" title="parts_submitted"><span class="displace">Parts Submitted</span></a></li> | ||
<li class="notebook"><a href="/Team:Greensboro-Austin/Notebook" title="notebook"><span class="displace">Notebook</span></a></li> | <li class="notebook"><a href="/Team:Greensboro-Austin/Notebook" title="notebook"><span class="displace">Notebook</span></a></li> | ||
| + | <li class="safety"><a href="/Team:Greensboro-Austin/Safety" title="safety"><span class="displace">Safety</span></a></li> | ||
<li class="attributions"><a href="/Team:Greensboro-Austin/Team#Attributions" title="attributions"><span class="displace">Attributions</span></a></li> | <li class="attributions"><a href="/Team:Greensboro-Austin/Team#Attributions" title="attributions"><span class="displace">Attributions</span></a></li> | ||
| - | |||
| - | |||
<!-- <li><img src="http://i.imgur.com/FBMrzYk.png" style="width:162.03px;height:567.6px"/></li> --> | <!-- <li><img src="http://i.imgur.com/FBMrzYk.png" style="width:162.03px;height:567.6px"/></li> --> | ||
| Line 39: | Line 38: | ||
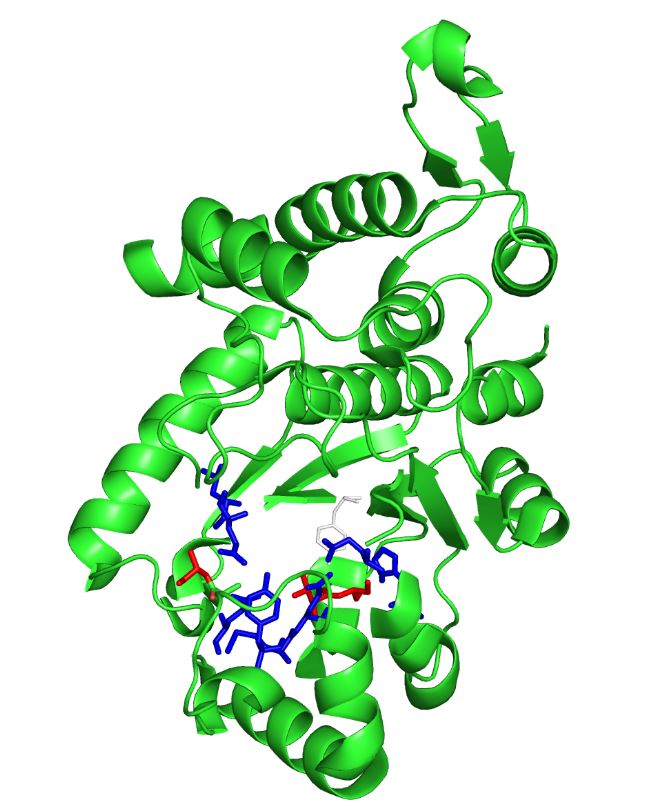
| - | The image on the left is the ''M. jannaschii'' synthetase mutated to incorporate 3-iodotyrosine by Sakamoto et. al. The blue and red residues make up the amino acid binding pocket, and the red residues are the ones that differ from the wild type ''M. jannaschii'' synthetase that incorporates tyrosine: they determine the specificity for | + | The image on the left is the ''M. jannaschii'' synthetase mutated to incorporate 3-iodotyrosine by Sakamoto et. al. The blue and red residues make up the amino acid binding pocket, and the red residues are the ones that differ from the wild type ''M. jannaschii'' synthetase that incorporates tyrosine: they determine the specificity for 3-iodotyrosine.[[#References|[3]]] The binding pocket is where mutations are made to incorporate different non-canonical amino acids such as 3,4-dihydroxy-L-phenylalanine (L-DOPA). |
| - | There are many non-canonical amino acids that have been incorporated through this expanded genetic code system including the | + | There are many non-canonical amino acids that have been incorporated through this expanded genetic code system including the 3-iodotyrosine synthetase used in the synthetase efficiency test below. We submitted the tRNA and synthetase pair that incorporate L-DOPA at the UAG codon to the parts registry as it is relevant to our mussel adhesion project; the synthetase binding pocket can be mutated by other teams to incorporate any of the other non-canonical amino acids.[[#References|[4]]] |
===Efficiency=== | ===Efficiency=== | ||
To test that the ''M. jannaschii'' synthetase and tRNA pair actually incorporate a non-canonical amino acid at the Amber stop codon, we designed a plasmid containing GFP with an Amber stop codon in place of the tyrosine codon at the active site (Tyr66). If translation stops at this site, then no GFP fluorescence can be seen; if an amino acid other than tyrosine is incorporated at this site, the emission wavelength of the fluorescence may be shifted. We tested this GFP plasmid in ''E. coli'' cells with the wild-type''M. jannaschii'' tyrosyl-tRNA and synthetase, in cells with the ''M. jannaschii'' tRNA and synthetase mutated to incorporate the non-canonical amino acid 3-iodotyrosine at UAG, in cells without an ''M. jannaschii'' synthetase, and compared their fluorescence to the expression of wild type GFP without an Amber stop codon in the active site. | To test that the ''M. jannaschii'' synthetase and tRNA pair actually incorporate a non-canonical amino acid at the Amber stop codon, we designed a plasmid containing GFP with an Amber stop codon in place of the tyrosine codon at the active site (Tyr66). If translation stops at this site, then no GFP fluorescence can be seen; if an amino acid other than tyrosine is incorporated at this site, the emission wavelength of the fluorescence may be shifted. We tested this GFP plasmid in ''E. coli'' cells with the wild-type''M. jannaschii'' tyrosyl-tRNA and synthetase, in cells with the ''M. jannaschii'' tRNA and synthetase mutated to incorporate the non-canonical amino acid 3-iodotyrosine at UAG, in cells without an ''M. jannaschii'' synthetase, and compared their fluorescence to the expression of wild type GFP without an Amber stop codon in the active site. | ||
| - | [[File:synthgraph.png|thumb|The red curve represents normal wild type GFP fluorescence without an Amber stop codon (Wild Type GFP), the blue curve labeled MjYRS represents fluorescence of the GFP plasmid with the amber stop codon at the active site of GFP in cells with the M. jannaschii tyrosine synthetase and tRNA (MjYRS), the green curve represents fluorescence of the same GFP plasmid in cells with the M. jannaschii synthetase and tRNA mutated to incorporate | + | [[File:synthgraph.png|thumb|The red curve represents normal wild type GFP fluorescence without an Amber stop codon (Wild Type GFP), the blue curve labeled MjYRS represents fluorescence of the GFP plasmid with the amber stop codon at the active site of GFP in cells with the M. jannaschii tyrosine synthetase and tRNA (MjYRS), the green curve represents fluorescence of the same GFP plasmid in cells with the M. jannaschii synthetase and tRNA mutated to incorporate 3-iodotyrosine (IodoY RS), and the turquoise curve represents fluorescence of the same GFP plasmid in cells without an M. jannaschii synthetase and tRNA pair (No ncAA RS).|500px|center]] |
| + | |||
| + | The mutated GFP with tyrosine incorporated at the UAG in the active site has the same intensity and wavelength of fluorescence as wild typre GFP which makes sense since their is no change in the amino acid sequence. The mutated GFP with 3-iodotyrosine incorporated at the active site has lower intensity showing that the ''M. jannaschii'' synthetase is not as efficient at incorporating 3-iodotyrosine, and the 10 nm shift in emission wavelength shows that 3-iodotyrosine is indeed being incorporated into and altering the fluorophore of GFP. All of the variants were carried out in RF0 ''E. coli'' cells except for the variant without an ''M. jannaschii'' synthetase which was done in normal BL21 ''E. coli'' because RF0 cells cannot survive without a tRNA and synthetase pair to code for an amino acid at the UAG Amber stop codon. | ||
| - | + | Our goal is to apply this technology to directly incorporate the non-canonical amino acid L-DOPA in our Mussel Adhesion Project, GluE.coli. | |
== References == | == References == | ||
Latest revision as of 03:51, 28 September 2013
Non-canonical amino acids
The "genetic code" refers to the rules by which RNA is translated into the proteins which perform most cellular functions. The genetic code is highly conserved throughout all organisms on earth, with 20 amino acids considered to be "canonical". However, many organisms encode non-canonical amino acids, or will modify residues after translation. Therefore, it is possible to transfer these exceptions to the code into organisms commonly used in molecular biology such as E. coli. This increases the toolbox of reactions and functions available when engineering new proteins.
One such recoding method is repurposing one of the stop codons in E. coli. There are three stop codons available in E. coli: amber, ochre and opal. The Amber stop codon, UAG, is used relatively rarely, and therefore presents an excellent target for amino acid recoding. In fact, this recoding already occurs in the Archaea [http://en.wikipedia.org/wiki/Methanocaldococcus_jannaschii Methanocaldococcus jannaschii]. M. jannashii has a tyrosyl-tRNA synthetase pair where the tRNA is charged with tyrosine and the anticodon loop recognizes the Amber stop codon. The active site of the orthogonal synthetase can be mutated to charge the tRNA with a non-canonical amino acid instead of tyrosine. [1]
Attempting to recode the Amber codon leads to several complications. There are several essential genes which terminate in the Amber stop codon, and which do not function properly if a non-canonical amino acid is used instead of translation stopping. Additionally, the release factor RF1 is responsible for terminating translation when the ribosome reaches an Amber stop codon, and will compete with the tRNA carrying the non-canonical amino acid. In order to fully recode the Amber codon, the strain RF0 [2] contains an F plasmid with all the essential genes normally ending in the amber codon mutated to end in another stop codon, and has RF1 knocked out to facilitate ncAA incorporation.
The image on the left is the M. jannaschii synthetase mutated to incorporate 3-iodotyrosine by Sakamoto et. al. The blue and red residues make up the amino acid binding pocket, and the red residues are the ones that differ from the wild type M. jannaschii synthetase that incorporates tyrosine: they determine the specificity for 3-iodotyrosine.[3] The binding pocket is where mutations are made to incorporate different non-canonical amino acids such as 3,4-dihydroxy-L-phenylalanine (L-DOPA).
There are many non-canonical amino acids that have been incorporated through this expanded genetic code system including the 3-iodotyrosine synthetase used in the synthetase efficiency test below. We submitted the tRNA and synthetase pair that incorporate L-DOPA at the UAG codon to the parts registry as it is relevant to our mussel adhesion project; the synthetase binding pocket can be mutated by other teams to incorporate any of the other non-canonical amino acids.[4]
Efficiency
To test that the M. jannaschii synthetase and tRNA pair actually incorporate a non-canonical amino acid at the Amber stop codon, we designed a plasmid containing GFP with an Amber stop codon in place of the tyrosine codon at the active site (Tyr66). If translation stops at this site, then no GFP fluorescence can be seen; if an amino acid other than tyrosine is incorporated at this site, the emission wavelength of the fluorescence may be shifted. We tested this GFP plasmid in E. coli cells with the wild-typeM. jannaschii tyrosyl-tRNA and synthetase, in cells with the M. jannaschii tRNA and synthetase mutated to incorporate the non-canonical amino acid 3-iodotyrosine at UAG, in cells without an M. jannaschii synthetase, and compared their fluorescence to the expression of wild type GFP without an Amber stop codon in the active site.
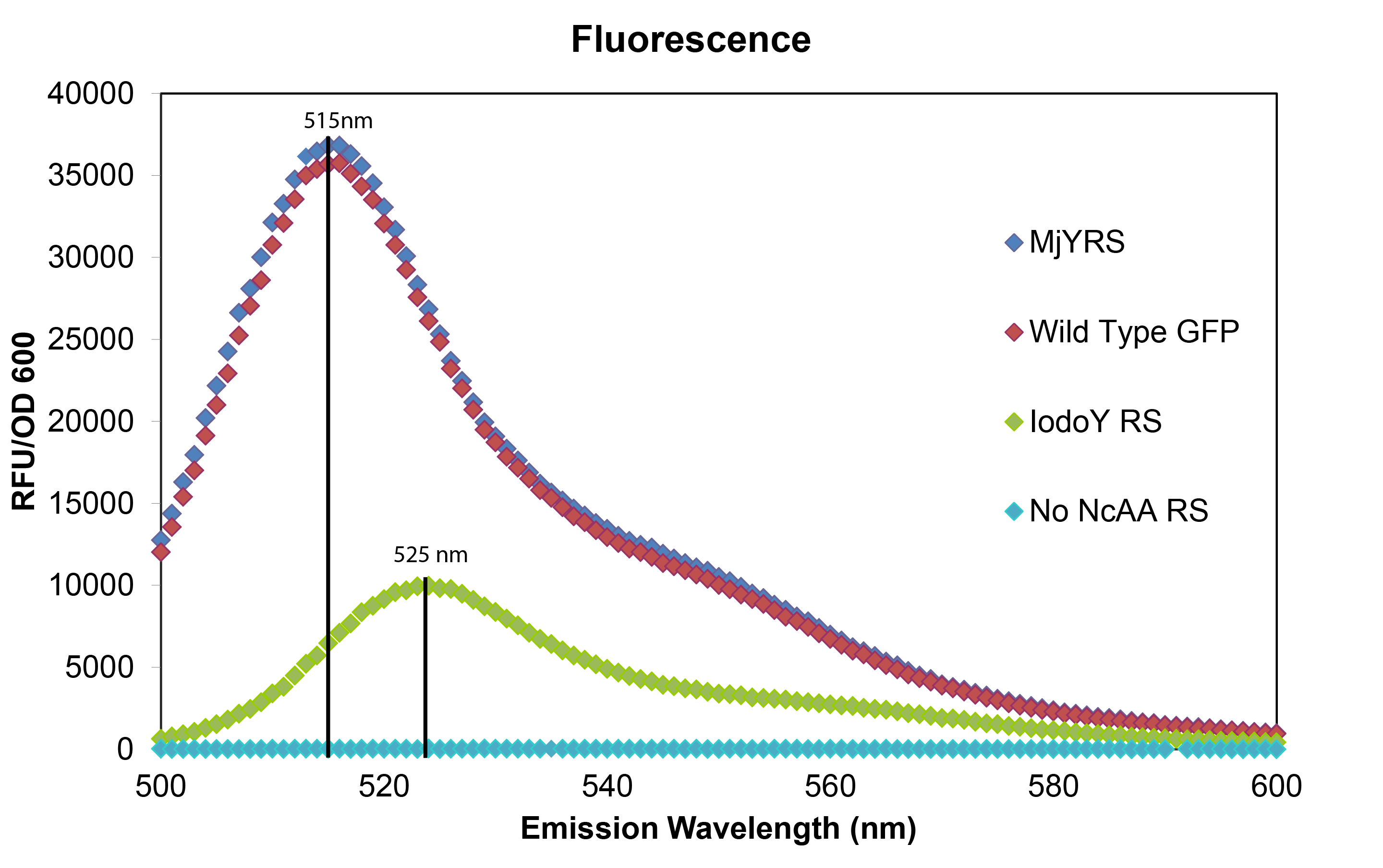
The mutated GFP with tyrosine incorporated at the UAG in the active site has the same intensity and wavelength of fluorescence as wild typre GFP which makes sense since their is no change in the amino acid sequence. The mutated GFP with 3-iodotyrosine incorporated at the active site has lower intensity showing that the M. jannaschii synthetase is not as efficient at incorporating 3-iodotyrosine, and the 10 nm shift in emission wavelength shows that 3-iodotyrosine is indeed being incorporated into and altering the fluorophore of GFP. All of the variants were carried out in RF0 E. coli cells except for the variant without an M. jannaschii synthetase which was done in normal BL21 E. coli because RF0 cells cannot survive without a tRNA and synthetase pair to code for an amino acid at the UAG Amber stop codon.
Our goal is to apply this technology to directly incorporate the non-canonical amino acid L-DOPA in our Mussel Adhesion Project, GluE.coli.
References
- Deiters A, and Schultz PG. In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorg Med Chem Lett. 2005 Mar 1;15(5):1521-4.Pubmed PMID: 15713420.
- Mukai T et. al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010 December; 38(22): 8188–8195. Pubmed PMID: 20702426
- Sakamoto et. al. Genetic Encoding of 3-Iodo-L-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography.Structure. 2009 Mar 11;17(3):335-44. doi: 10.1016/j.str.2009.01.008. PubMed PMID: 19278648.
- Liu CC, Schultz PG. Adding New Chemistries to the Genetic Code. Annu Rev Biochem. 2010;79:413-44. doi: 10.1146/annurev.biochem.052308.105824. PubMed PMID: 20307192.
- Wang L, et al. Expanding the Genetic Code of Escherichia coli. Science 20 April 2001: 292 (5516), 498-500.
 "
"