Team:NYMU-Taipei/Modeling/MainParts
From 2013.igem.org
| (64 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NYMU-Taipei/Header}} | {{:Team:NYMU-Taipei/Header}} | ||
| + | =Main parts= | ||
| + | ==Backgrounds== | ||
| - | + | '''Circuit regulators''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | • ptet regulated promoter (ptet) : when ptet exists, it will bind to ptet regulated promoter (ptet) and represses the promoter. | |
| - | + | • pLux/cI hybrid promoter (plux/cI) : when luxR/AHL exists, it will open pLux/cI hybrid promoter (plux/cI), while cI will repress the promoter. What’s more, plux/cI is cI-dominant, which means when cI exists, the hybrid promoter will be repressed whether luxR/AHL exists or not. | |
| - | + | ||
| - | + | '''Circuit regulation:''' | |
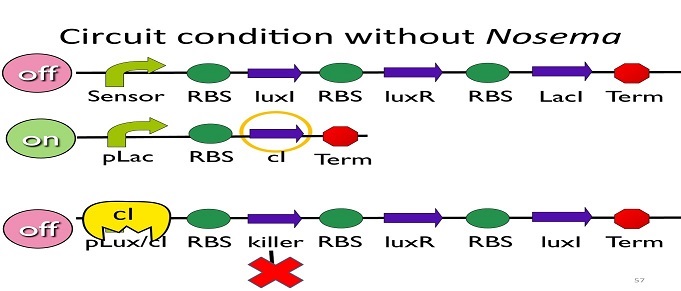
| - | + | • Circuit condition without ''Nosema'': | |
| + | [[Image:NYMU_ Circuit condition without Nosema.png|center]] | ||
| - | + | Without ''Nosema ceranae'', the first circuit will not open, and thus, no ptet will be produced. Since there is no ptet binding to ptet, ptet will not be repressed, which means cI will be produced. After that, cI will bind to pLux/cI hybrid promoter, leading the third circuit to be off. | |
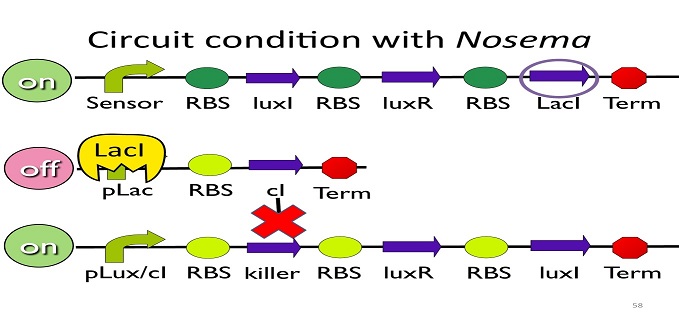
| - | + | • Circuit condition with'' Nosema'': | |
| - | + | [[Image:NYMU_ Circuit condition with Nosema.png|center]] | |
| - | + | When ''Nosema ceranae'' infects the bee, bee’s immune system will be activated, leading to the concentration of ROS (reactive oxygen species) to be high. In response, Bee. coli will enhance the production of oxyR, which forms a complex with ROS and binds to transcription binding site ahead of AhpCp (an oxyR-activated promoter; namely, the sensor), leading to the first circuit to be on. | |
| - | + | Since the first circuit is on, the downstream ptet gene will be produced and bind to ptet, leading to the second circuit to be off. | |
| - | + | ||
| - | + | After the second circuit is closed, no cI is produced, leading to the third circuit to be on (no repressor at all). And then, kill protein will be produced and kill ''Nosema Ceranae''. | |
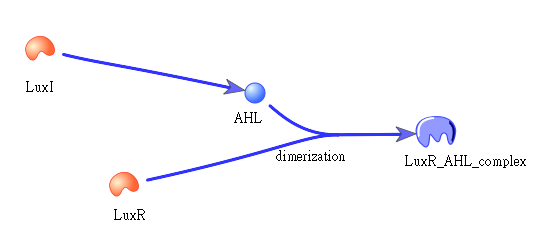
| - | + | • Positive feedback: | |
| - | + | [[Image:NYMU_positive feedback.png|center]] | |
| - | + | Besides the LuxI and LuxR(which then form a complex called luxR/AHL) produced by first circuit, the third circuit will also produce LuxI and LuxR, which acts as a positive feedback and strongly enhances the production of killer protein. | |
| - | + | The picture shows how LuxI will transfer to AHL, which then bind to LuxR to form a dimer: | |
| - | + | ||
| - | + | [[Image:NYMU_luxr ahl dimer.png|center]] | |
| - | + | ||
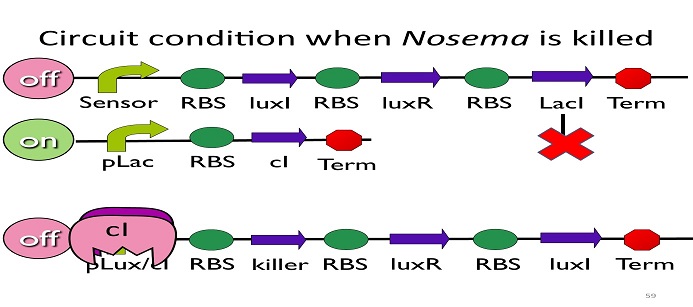
| - | It is assumed that AHL is abundant and thus the formation of AHL2LuxR is determined only by the concentration of LuxR; CI’s mechanism of binding to the promoter is assumed to act as hill effect form; kill protein will sustain a period of time before it is degraded naturally without any other types of | + | • Circuit condition when ''Nosema'' is killed: |
| + | [[Image:NYMU_ Circuit condition when Nosema is killed.png|center]] | ||
| + | |||
| + | After ''Nosema'' is killed, the sensor will be off and no ptet will be produced. | ||
| + | |||
| + | Without ptet, the ptet will not be repressed, and the second circuit will be on, leading to the production of cI. After cI binds to pLux/cI hybrid promoter, the third circuit to be off. After that, the overall circuit will switch to the beginning stage when there is no'' Nosema''. | ||
| + | |||
| + | ==Objectives== | ||
| + | |||
| + | 1. To know how much time it needs from sensing to killing the ''Nosema'' after infection | ||
| + | |||
| + | 2. To know whether the pathway is effective or not | ||
| + | |||
| + | 3. To know the range of kill protein concentration (from the minimal concentration which Is effective to kill ''Nosema'' to the maximal concentration which will not do harm to the bees) | ||
| + | |||
| + | |||
| + | ==System== | ||
| + | |||
| + | It is assumed that AHL is abundant and thus the formation of AHL2LuxR complex is determined only by the concentration of LuxR; CI’s mechanism of binding to the promoter is assumed to act as hill effect form; kill protein will sustain a period of time before it is degraded naturally without any other types of decomposition. | ||
| + | |||
| + | '''Equation1''' | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[ | + | \frac{d[mRNA_C_I]}{dt}= \frac{1-[ptet]^nptet}{ Kdptet^nptet+ [ptet]^nptet} |
| + | |||
| + | \times PoPSconstitutive\times\frac{N}{V} | ||
| + | |||
| + | -KdegmRNA\times [mRNA_C_I] | ||
</div> | </div> | ||
</html> | </html> | ||
| - | + | Kdptet = dissociation constant of CI | |
| - | + | nptet = Hill coefficient of ptet | |
| - | + | PoPSptet = promoter strength of ptet | |
kdegmRNA = degrading constant of sensor promoter mRNA | kdegmRNA = degrading constant of sensor promoter mRNA | ||
| Line 60: | Line 78: | ||
V = volume of a cell | V = volume of a cell | ||
| - | '''The aim of the equation is to | + | ''' The aim of the equation is to know mRNA_C_I production rate and when it can reach the level to translate the desired concentration.''' |
| - | + | [[read more1]] | |
| + | '''Equation2''' | ||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[ | + | \frac{d[mRNA_L_a_c_I]}{dt}= \frac{1-[ROSoxyR]^nROSoxyR }{ KdROSoxyR ^nROSoxyR + [ROSoxyR]^nROSoxyR} PoPSAhpCp\times\frac{N}{V}-KdegmRNA[mRNA_L_a_c_I] |
</div> | </div> | ||
</html> | </html> | ||
| Line 74: | Line 93: | ||
nROSoxyR = Hill coefficient of ROSoxyR | nROSoxyR = Hill coefficient of ROSoxyR | ||
| - | + | PoPSAhpCp = promoter strength of AhpCp | |
kdegmRNA = degrading constant of sensor promoter mRNA | kdegmRNA = degrading constant of sensor promoter mRNA | ||
| Line 82: | Line 101: | ||
V = volume of a cell | V = volume of a cell | ||
| - | '''The aim of the equation is to know how | + | ''' The aim of the equation is to know how AhpCp promoter strength (in PoPS) influences the production of ptet.''' |
| + | |||
| + | [[read more2]] | ||
| + | |||
| + | '''Equation3:''' | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[ | + | \frac{d[mRNA_O_x_y_R]}{dt}=PoPSconstitutive\times\frac{N}{V}-KdegmRNA\times [mRNAOxyR]</div> |
| - | </div> | + | |
</html> | </html> | ||
| Line 99: | Line 120: | ||
kdegmRNA = degrading constant of sensor promoter mRNA | kdegmRNA = degrading constant of sensor promoter mRNA | ||
| - | '''The aim of the equation is to know the production rate ofmRNAoxyR, and choose the proper constitutive promoter for boosting OxyR's concentration.''' | + | ''' The aim of the equation is to know the production rate ofmRNAoxyR, and choose the proper constitutive promoter for boosting OxyR's concentration.''' |
| - | + | [[read more3]] | |
| - | <html> | + | |
| - | <div lang="latex" class="equation"> | + | '''Equation4:''' |
| - | + | ||
| - | </div> | + | <html> <div lang="latex" class="equation"> |
| - | </html> | + | \frac{d[mRNA_L_u_x_I]}{dt}= \frac{1-[ROSoxyR]^n ROSoxyR }{ Kd ROSoxyR ^n ROSoxyR + [ROSoxyR]^n ROSoxyR I} PoPSAhpCp\times\frac{N}{V}+\frac{1-[ LuxRAHL]^nLuxRAHL}{ KdLuxRAHL ^nLuxRAHL + [LuxRAHL]^nLuxRAHL}\times\frac{1-{1-[CI leak]^nCI}}{ KdCI^nCI+[CI]^nCI}\times PoPSLuxCI\times\frac{N}{V}-KdegmRNA[mRNALuxI] |
| + | </div> </html> | ||
KdROSoxyR = dissociation constant ofROSoxyR | KdROSoxyR = dissociation constant ofROSoxyR | ||
| Line 112: | Line 134: | ||
nROSoxyR = Hill coefficient of ROSoxyR | nROSoxyR = Hill coefficient of ROSoxyR | ||
| - | + | PoPSAhpCp = promoter strength of AhpCp | |
KdLuxRAHL = dissociation constant ofLuxRAHL | KdLuxRAHL = dissociation constant ofLuxRAHL | ||
| Line 118: | Line 140: | ||
nLuxRAHL = Hill coefficient of LuxRAHL | nLuxRAHL = Hill coefficient of LuxRAHL | ||
| - | KdcI = dissociation constant | + | KdcI = dissociation constant of CI |
| - | ncI = Hill coefficient | + | ncI = Hill coefficient of CI |
PoPSLuxcI = promoter strength of LuxcI hybrid promoter | PoPSLuxcI = promoter strength of LuxcI hybrid promoter | ||
| - | kdegmRNA = degrading constant of sensor promoter | + | kdegmRNA = degrading constant of sensor promoter Mrna |
N = number of plasmid in a single cell | N = number of plasmid in a single cell | ||
| Line 130: | Line 152: | ||
V = volume of a cell | V = volume of a cell | ||
| - | '''The aim of the equation is to know how | + | ''' The aim of the equation is to know how AhpCp promoter strength (in PoPS) influences the production of LuxI. ''' |
| - | + | [[read more4]] | |
| - | <html> | + | |
| - | <div lang="latex" class="equation"> | + | '''Equation5:''' |
| - | + | ||
| - | </div> | + | <html> <div lang="latex" class="equation"> |
| - | </html> | + | \frac{d[mRNA_L_u_x_R]}{dt}= \frac{1-[ROSoxyR]^n ROSoxyR }{ Kd ROSoxyR ^n ROSoxyR + [ROSoxyR]^n ROSoxyR I} \times PoPSAhpCp\times\frac{N}{V}+\frac{1-[ LuxRAHL]^nLuxRAHL}{ KdLuxRAHL ^nLuxRAHL + [LuxRAHL]^nLuxRAHL}\times\frac{1-{1-[CI leak]^nCI}}{ KdCI^nCI+[CI]^nCI}\times PoPSLuxCI\times\frac{N}{V}-KdegmRNA[mRNALuxR] |
| + | </div> </html> | ||
KdROSoxyR = dissociation constant ofROSoxyR | KdROSoxyR = dissociation constant ofROSoxyR | ||
| Line 143: | Line 166: | ||
nROSoxyR = Hill coefficient of ROSoxyR | nROSoxyR = Hill coefficient of ROSoxyR | ||
| - | + | PoPSAhpCp = promoter strength of AhpCp | |
KdLuxRAHL = dissociation constant ofLuxRAHL | KdLuxRAHL = dissociation constant ofLuxRAHL | ||
| Line 149: | Line 172: | ||
nLuxRAHL = Hill coefficient of LuxRAHL | nLuxRAHL = Hill coefficient of LuxRAHL | ||
| - | KdcI = dissociation constant | + | KdcI = dissociation constant of CI |
| - | ncI = Hill coefficient | + | ncI = Hill coefficient of CI |
PoPSLuxcI = promoter strength of LuxcI hybrid promoter | PoPSLuxcI = promoter strength of LuxcI hybrid promoter | ||
| - | kdegmRNA = degrading constant of sensor promoter | + | kdegmRNA = degrading constant of sensor promoter Mrna |
N = number of plasmid in a single cell | N = number of plasmid in a single cell | ||
| Line 161: | Line 184: | ||
V = volume of a cell | V = volume of a cell | ||
| - | '''The aim of the equation is to know how | + | ''' The aim of the equation is to know how AhpCp promoter strength (in PoPS) influences the production of LuxR. ''' |
| - | + | [[read more5]] | |
| - | <html> | + | |
| - | <div lang="latex" class="equation"> | + | '''Equation6''' |
| - | + | ||
| - | </div> | + | <html> <div lang="latex" class="equation"> |
| - | </html> | + | \frac{d[mRNA_k_i_l_l]}{dt}= \frac{1-[ LuxRAHL]^nLuxRAHL}{ KdLuxRAHL ^nLuxRAHL + [LuxRAHL]^nLuxRAHL}\times\frac{1-{1-[CI leak]^nCI}}{ KdCI^nCI+[CI]^nCI}\times PoPSLuxCI\times\frac{N}{V}-KdegmRNA[mRNA_k_i_l_l] |
| + | </div> </html> | ||
KdLuxRAHL = dissociation constant ofLuxRAHL | KdLuxRAHL = dissociation constant ofLuxRAHL | ||
| Line 174: | Line 198: | ||
nLuxRAHL = Hill coefficient of LuxRAHL | nLuxRAHL = Hill coefficient of LuxRAHL | ||
| - | KdcI = dissociation constant | + | KdcI = dissociation constant of CI |
| - | ncI = Hill coefficient | + | ncI = Hill coefficient of CI |
PoPSLuxcI = promoter strength of LuxcI hybrid promoter | PoPSLuxcI = promoter strength of LuxcI hybrid promoter | ||
| Line 186: | Line 210: | ||
V = volume of a cell | V = volume of a cell | ||
| - | '''The aim of the equation is to | + | '''The aim of the equation is to know mRNA of kill protein production rate and when it can reach the level of the desired concentration. ''' |
| - | + | [[read more6]] | |
| - | <html> | + | |
| - | <div lang="latex" class="equation"> | + | [[to learn more about how we characterize pLuxCI, click here]] |
| - | + | ||
| - | </div> | + | |
| - | </html> | + | '''Equation7''' |
| + | |||
| + | <html> <div lang="latex" class="equation"> | ||
| + | \frac{d[mRNA_t_7]}{dt}= \frac{1-[ROSoxyR]^n ROSoxyR }{ Kd ROSoxyR ^n ROSoxyR + [ROSoxyR]^nROSoxyR }\timesPoPSAhpCp\times\frac{N}{V}\times {a}+ \frac{1-[T7]^nT7}{ Kd T7^nT7 + [T7]^nT7}\times{PoPST7}\times\ frac{N}{V}-KdegmRNA\times [mRNA_T_7] | ||
| + | </div> </html> | ||
KdROSoxyR = dissociation constant ofROSoxyR | KdROSoxyR = dissociation constant ofROSoxyR | ||
| Line 199: | Line 227: | ||
nROSoxyR = Hill coefficient of ROSoxyR | nROSoxyR = Hill coefficient of ROSoxyR | ||
| - | + | PoPSAhpCp = promoter strength of AhpCp | |
KdT7 = dissociation constant of T7 | KdT7 = dissociation constant of T7 | ||
| Line 213: | Line 241: | ||
V = volume of a cell | V = volume of a cell | ||
| - | '''The aim of the equation is to | + | '''The aim of the equation is to know mRNA of T7 polymerase production rate and when it can reach the level to translate enough T7 polymerase. ''' |
| + | |||
| + | [[read more7]] | ||
| + | |||
| + | '''Equation8:''' | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[ | + | \frac{d[mRNA_P_D_C]}{dt}=\frac{ [T7]^{nT7} }{ KdT7^{nT7}+[T7]^{nY7} }\times{PoPST7}\times\frac{N}{V}-KdegmRNA\times[mRNAPDC] |
</div> | </div> | ||
</html> | </html> | ||
| - | + | <br> | |
KdT7 = dissociation constant of T7 | KdT7 = dissociation constant of T7 | ||
| Line 228: | Line 259: | ||
PoPST7 = promoter strength of T7 promoter | PoPST7 = promoter strength of T7 promoter | ||
| - | + | KdegmRNA = degrading constant of sensor promoter mRNA | |
| - | N = number of plasmid in a single cell | + | N = number of plasmid in a single cell |
V = volume of a cell | V = volume of a cell | ||
| - | '''The aim of the equation is to knowmRNA of enzyme PDC production rate and when it can reach the level to translate enough PDC.''' | + | '''The aim of the equation is to knowmRNA of enzyme PDC production rate and when it can reach the level to translate enough PDC. ''' |
| + | |||
| + | [[read more8]] | ||
| + | |||
| + | '''Equation9:''' | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[ | + | \frac{d[mRNA_A_D_H]}{dt}=\frac{ [T7]^{nT7} }{ KdT7^{nT7}+[T7]^{nY7} }\times{PoPST7}\times\frac{N}{V}-KdegmRNA\times[mRNAADH] |
</div> | </div> | ||
</html> | </html> | ||
| Line 255: | Line 289: | ||
V = volume of a cell | V = volume of a cell | ||
| - | '''The aim of the equation is to knowmRNA of enzyme ADH production rate and when it can reach the level to translate enough ADH.''' | + | '''The aim of the equation is to knowmRNA of enzyme ADH production rate and when it can reach the level to translate enough ADH. ''' |
| + | |||
| + | [[read more9]] | ||
| + | |||
| + | |||
| + | '''Equation10 ''' | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[CI]}{dt}=RBS\times[ | + | \frac{d[CI]}{dt}={RBS}\times{[mRNA_C_I]} -KdegCI\times{[CI]} |
</div> | </div> | ||
</html> | </html> | ||
| Line 268: | Line 306: | ||
KdegCI = degrading constant of CI | KdegCI = degrading constant of CI | ||
| - | '''The aim of the equation is to know the production rate of CI and when it can reach the concentration to repress LuxCI hybrid promoter.''' | + | '''The aim of the equation is to know the production rate of CI and when it can reach the concentration to repress LuxCI hybrid promoter. ''' |
| + | |||
| + | [[read more10]] | ||
| + | |||
| + | '''Equation11''' | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{[ | + | \frac{d[ptet]}{dt}={RBS}\times{[mRNA_L_a_c_I]} - {Kdeg ptet} \times{[ptet]} |
</div> | </div> | ||
</html> | </html> | ||
| Line 279: | Line 320: | ||
RBS = binding site strength | RBS = binding site strength | ||
| - | + | Kdegptet = degrading constant of ptet | |
| - | '''The aim of the equation is to | + | '''The aim of the equation is to known ptet production rate and when it can reach the concentration to repress promoter ptet.''' |
| + | |||
| + | [[read more11]] | ||
| + | |||
| + | '''Equation12'' | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{[ | + | \frac{d[LuxI]}{dt}={RBS}\times{[mRNA_L_u_x_I]} -KAHL\times{[LuxI]} |
| + | -Kdeg {[LuxI]} \times{[LuxI]} | ||
</div> | </div> | ||
</html> | </html> | ||
| Line 292: | Line 337: | ||
RBS = binding site strength | RBS = binding site strength | ||
| - | KAHL = | + | KAHL = LuxI to AHL rate constant |
KdegLuxI = degrading constant of LuxI | KdegLuxI = degrading constant of LuxI | ||
| - | '''The aim of the equation is to know the production rate of LuxI concerning the LuxIAHL reaction and LuxI degrading.''' | + | '''The aim of the equation is to know the production rate of LuxI concerning the LuxIAHL reaction and LuxI degrading. ''' |
| + | |||
| + | [[read more12]] | ||
| + | |||
| + | '''Equation13'' | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[LuxR]}{dt}=RBS\times[ | + | \frac{d[LuxR]}{dt}={RBS}\times{[mRNA_L_u_x_R]} – {KmAHL}\times{[AHL]} |
| + | - {Kdeg LuxR} \times{[LuxR]} \times {KmAHL}\{times[AHL]^{2}} \times{[LuxR]}- KdegAHL\times{[AHL]} | ||
</div> | </div> | ||
</html> | </html> | ||
| Line 307: | Line 356: | ||
RBS = binding site strength | RBS = binding site strength | ||
| - | KmAHL = | + | KmAHL =AHL to LuxI rate constant |
KdegLuxR= degrading constant of LuxR | KdegLuxR= degrading constant of LuxR | ||
| - | KonAHL = 2AHL + | + | KonAHL = 2AHL + LuxR to (AHL)2/LuxR complex rate constant |
KdegAHL = degrading constant of AHL | KdegAHL = degrading constant of AHL | ||
| - | '''The aim of the equation is to know the production rate of LuxR concerning the 2AHL + LuxR | + | '''The aim of the equation is to know the production rate of LuxR concerning the 2AHL + LuxR to (AHL)2/LuxR reaction , AHL to LuxI reaction, and LuxR, AHL degrading. ''' |
| + | |||
| + | [[read more13]] | ||
| + | |||
| + | '''Equation14:'' | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[AHL]}{dt}=KAHL\times[LuxI]+2\ | + | \frac{d[AHL]}{dt}={KAHL}\times{[LuxI]}+2\times KoffAHL \times [AHLLuxR] - 2\times KonAHL \times {[AHL]}^{2}\times {[LuxR]} - KdegAHL\times{[AHL]} |
</div> | </div> | ||
</html> | </html> | ||
| - | KAHL = | + | KAHL = LuxI to AHL rate constant |
| - | KonAHL = 2AHL + | + | KonAHL = 2AHL + LuxR to (AHL)^2/LuxR complex rate constant |
| - | KoffAHL = (AHL)2/LuxR | + | KoffAHL = (AHL)2/LuxR complex to 2AHL + LuxR rate constant |
KdegAHL = degrading constant of AHL | KdegAHL = degrading constant of AHL | ||
| - | '''The aim of the equation is to know the production rate of AHL concerning the | + | '''The aim of the equation is to know the production rate of AHL concerning the LuxIAHL reaction, 2AHL + LuxR to (AHL)2/LuxR reaction, (AHL)2/LuxR complex to 2AHL + LuxR reaction and AHL degrading. ''' |
| + | |||
| + | [[read more14]] | ||
| + | |||
| + | |||
| + | '''Equation17''' | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[ | + | \frac{d[ADH]}{dt}=RBS\times{PoPST7effect}\times\frac{N}{V}-{KdegADH}\times{[ADH]} |
</div> | </div> | ||
</html> | </html> | ||
| + | <br> | ||
RBS = binding site strength | RBS = binding site strength | ||
| - | + | PoPST7 = promoter strength of T7 promoter | |
| - | + | N = number of plasmid in a single cell | |
| - | = | + | V = volume of a cell |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | KdegADH = degrading constant of ADH (Acetaldehyde) | |
| - | + | '''The aim of the equation is to know ADH production rate and when it can reach the concentration of ethanol pathway equilibrium.''' | |
| - | + | [[read more17]] | |
| - | + | '''Equation18''' | |
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[ | + | \frac{d[AHL]}{dt}=\times KonAHL \times [AHL]^{2}\times{[LuxR]}- {KoffAHL} \times {[AHLLuxR]} - KdegAHL\times{[AHL]} |
</div> | </div> | ||
</html> | </html> | ||
| - | + | KAHL = LuxI to AHL rate constant | |
| - | + | KonAHL = 2AHL + LuxR to (AHL)2/LuxR complex rate constant | |
| - | + | KoffAHL = (AHL)2/LuxR complex to 2AHL + LuxR rate constant | |
| - | = | + | KdegAHL = degrading constant of AHL |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | '''The aim of the equation is to know the production rate of AHL concerning the LuxIAHL reaction, 2AHL + LuxR to (AHL)2/LuxR reaction, (AHL)2/LuxR complex to 2AHL + LuxR reaction and AHL degrading. ''' | |
| - | + | [[read more18]] | |
| - | ''' | + | '''Equation19''' |
| - | + | ||
| - | |||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[kill]}{dt}=RBS\times[ | + | \frac{d[kill]}{dt}={RBS}\times{[mRNA_k_i_l_l]} –Kdegkill\times{[kill]} |
</div> | </div> | ||
</html> | </html> | ||
| Line 398: | Line 443: | ||
Kdegkill = degrading constant of kill | Kdegkill = degrading constant of kill | ||
| - | '''The aim of the equation is to knowkiller protein production rate and when it can reach the effective concentration to | + | '''The aim of the equation is to knowkiller protein production rate and when it can reach the effective concentration to kill ''Nosema''. ''' |
| - | + | [[read more19]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | + | ==Results== | |
| + | [[File:NYMU_defensin normal.png]] | ||
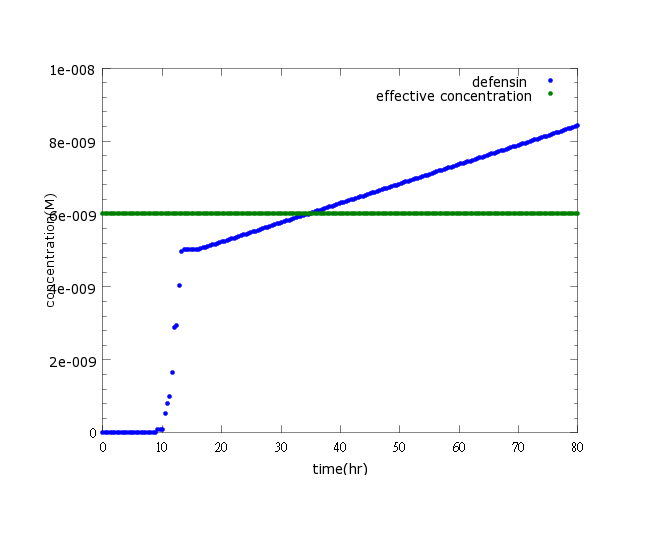
| - | + | Figure1: This picture shows kill protein concentration to time under pLux/CI hybrid promoter’s control and LuxR, LuxI’s positive feedback. The result indicates that kill protein concentration will reach its effective level and kill ''Nosema'' in time (36 hours after ''Nosema'' infection) | |
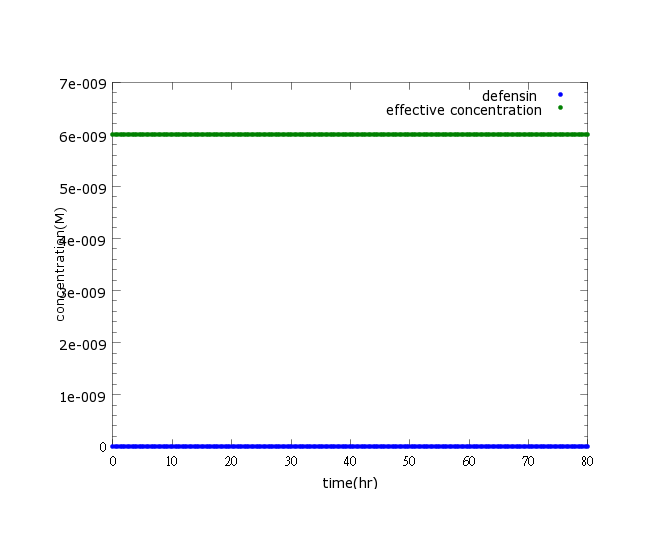
| - | + | [[File:NYMU_defensin witout ROS.png]] | |
| - | ''' | + | Figure2: This picture is a control group model. It shows that without ''Nosema'' infection (ROS concentration=0), ethanol production is low enough to be ignored. |
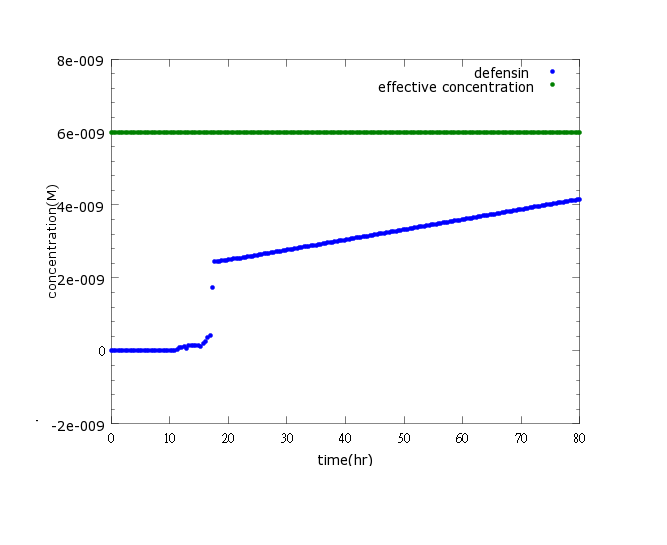
| - | + | [[File:NYMU_defensin only CI.png]] | |
| - | + | Figure3: This is the picture of kill protein concentration to time under promoter CI’s control only(control group)without pLux/CI's regulation.The result shows that kill protein's full expression is too low to kill ''Nosema''. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ==Discussion== | |
| - | + | ||
| - | + | Comparing to the ROS-level control group where ROS remains low when there’s no ''Nosema'' infection, kill protein production is highly boasted upon sensing ''Nosema''. | |
| - | + | ||
| - | + | The result indicates that kill protein concentration will reach its effective level and kill ''Nosema'' in time (36 hours after sensing ''Nosema'' infection, within the 5-day deadline when'' Nosema'' spores proliferates and the bee become incurable and infectious to other bees), at the given ROS changes and the enhanced OxyR expression [[(to learn more about our sensor model, click here)]]. | |
| - | + | ||
| - | + | ==Circuit design and improvement== | |
| + | |||
| + | Upon using pCI to negatively regulate kill protein production, results show that kill protein's full expression while no CI present is too low to kill ''Nosema''. To solve this problem we subsidized CI promoter into pLux/CI promoter for additional positive feedback to boast kill protein’s production. As the modeling results regarding the modified circuit shown, the consequences are positive. [[(click here to learn more about our circuit)]] | ||
| + | |||
| + | ==Parameters:== | ||
| + | {|class="wikitable" !Model!!Parameter!!Description!!Value!!Unit!!Reference | ||
| + | ! style="text-align: center;"|Model | ||
| + | ! style="text-align: center;"|Parameter | ||
| + | ! style="text-align: center;"|Description | ||
| + | ! style="text-align: center;"|Value | ||
| + | ! style="text-align: center;"|Unit | ||
| + | ! style="text-align: center;"|Reference | ||
| + | |- | ||
| + | |rowspan=5 style="text-align: center;"|'''Sensor''' | ||
| + | | style="text-align: center;"|KdegOxyR | ||
| + | |OxyR degrading rate | ||
| + | | style="text-align: center;"|10<sup>7</sup> | ||
| + | | style="text-align: center;"|M<sup>-1</sup> x min<sup>-1</sup> | ||
| + | |rowspan=3|Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol—disulfide status | ||
| + | |- | ||
| + | | style="text-align: center;"|KdOxyR* | ||
| + | |Dissociation constant of OxyR* | ||
| + | | style="text-align: center;"|10<sup>-7.33</sup> | ||
| + | | style="text-align: center;"|M | ||
| + | |- | ||
| + | | style="text-align: center;"|KOxyR | ||
| + | |OxyR producing rate constant | ||
| + | | style="text-align: center;"|5.012 x 10<sup>14</sup> | ||
| + | | style="text-align: center;"|X | ||
| + | |- | ||
| + | | style="text-align: center;"|nOxyR* | ||
| + | |Hill coefficient of OxyR* | ||
| + | |Ranging from 0.75~3.5,depends on what kind of ROS it react with | ||
| + | | style="text-align: center;"|X | ||
| + | |OxyR: A Molecular Code for Redox-Related Signaling | ||
| + | |- | ||
| + | | style="text-align: center;"|N | ||
| + | |Copy number | ||
| + | |887(cells containing 25% plasmid bearing cells) | ||
| + | |Single piece | ||
| + | | | ||
| + | #Escherichia coli Plasmid Copy Number Assay | ||
| + | #Improved determination of plasmid copy number using quantitative real-time PCR for monitoring fermentation processes | ||
| + | |- | ||
| + | | rowspan="3" style="text-align: center;" | Ethanol | ||
| + | |||
| + | |||
| + | | colspan="1" style="text-align: center;" |pyruvate | ||
| + | | colspan="1" style="text-align: center;" | Initial concentration of pyruvate in MG1655 | ||
| + | | colspan="1" style="text-align: center;" | 1.18 x 2 | ||
| + | | colspan="1" style="text-align: center;" | g/L | ||
| + | | colspan="1" style="text-align: center;" |Expression of pyruvate carboxylase enhances succinate production in Escherichia coli without affecting glucose uptake | ||
| + | |- | ||
| + | |style="text-align: center;" | nPDC | ||
| + | | Hill coefficient of PDC | ||
| + | | style="text-align: center;" |2.1 | ||
| + | | style="text-align: center;" | - | ||
| + | |Purification, characterization and cDNA sequencing of pyruvate decarboxylase Zygosaccharomyces biporus | ||
| + | |- | ||
| + | |style="text-align: center;" |nADH||Hill coefficient of ADH|| style="text-align: center;" |at high pH values, 30◦C, n=1; at low temperature, n=3 | ||
| + | |style="text-align: center;" | - | ||
| + | |Evidence for co-operativity in coenzyme binding to tetrameric Sulfolobus | ||
| + | solfataricus alcohol dehydrogenase and its structural basis: fluorescence, | ||
| + | kinetic and structural studies of the wild-type enzyme and non-co-operative | ||
| + | N249Y mutant | ||
| + | |||
| + | |- | ||
| + | |- | ||
| + | | rowspan="6" | SEIR(exponential) | ||
| + | |||
| + | |||
| + | | colspan="1" style="text-align: center;" | b | ||
| + | | colspan="1" style="text-align: center;" | Infection rate constant of ''Nosema'' ceranae to the suspected | ||
| + | | colspan="1" style="text-align: center;" | 24/75 | ||
| + | | colspan="1" style="text-align: center;" | Period(days)-1 | ||
| + | | colspan="1" style="text-align: center;" | | ||
| + | |- | ||
| + | |style="text-align: center;" | r1 | ||
| + | | Infection rate constant of K12 to the suspected | ||
| + | | style="text-align: center;" |3/20 | ||
| + | | Period(days)-1 | ||
| + | | 1. Environment protection administration executive yuan of R.O.C Medical bacteriology of J.A.T | ||
| + | |||
| + | |- | ||
| + | |style="text-align: center;" |r2||Infection rate constant of K12 to the latent|| style="text-align: center;" |3/20 ||Period(days)-1||rowspan="2" | 2. Environment protection administration executive yuan of R.O.C.<br> 3. Medical bacteriology of J.A.T | ||
| + | |||
| + | |- | ||
| + | |style="text-align: center;" | e ||rate of the latent turns infectious||style="text-align: center;" |1/4||Period(days)-1 | ||
| + | |- | ||
| + | |style="text-align: center;" | u || Death rate of the infected || style="text-align: center;" |1/8||Period(days)-1|| | ||
| + | |- | ||
| + | |style="text-align: center;" | k || Rate of intaking capsule ||style="text-align: center;" |24/11||Period(days)-1|| | ||
| + | |- | ||
| + | | rowspan="4" |SEIR(exponential&linear) | ||
| + | | colspan="1" style="text-align: center;" | S | ||
| + | | colspan="1" style="text-align: center;" | x(1) | ||
| + | | colspan="1" style="text-align: center;" | Amount of total population | ||
| + | | colspan="1" style="text-align: center;" | | ||
| + | | colspan="1" style="text-align: center;" | | ||
| + | |- | ||
| + | |style="text-align: center;" | E | ||
| + | | style="text-align: center;" |x(2) | ||
| + | |style="text-align: center;" | Amount of suspected individuals | ||
| + | | | ||
| + | | | ||
| + | |||
| + | |- | ||
| + | |style="text-align: center;" |I||style="text-align: center;" |x(3)|| style="text-align: center;" |Amount of individuals in the latent period || || | ||
| + | |||
| + | |- | ||
| + | |style="text-align: center;" | R ||style="text-align: center;" |x(4)||style="text-align: center;" |Amount of infected individuals|| || | ||
| + | |- | ||
| + | |||
| + | |} | ||
{{:Team:NYMU-Taipei/Footer}} | {{:Team:NYMU-Taipei/Footer}} | ||
Latest revision as of 02:40, 29 October 2013


Contents |
Main parts
Backgrounds
Circuit regulators
• ptet regulated promoter (ptet) : when ptet exists, it will bind to ptet regulated promoter (ptet) and represses the promoter.
• pLux/cI hybrid promoter (plux/cI) : when luxR/AHL exists, it will open pLux/cI hybrid promoter (plux/cI), while cI will repress the promoter. What’s more, plux/cI is cI-dominant, which means when cI exists, the hybrid promoter will be repressed whether luxR/AHL exists or not.
Circuit regulation:
• Circuit condition without Nosema:
Without Nosema ceranae, the first circuit will not open, and thus, no ptet will be produced. Since there is no ptet binding to ptet, ptet will not be repressed, which means cI will be produced. After that, cI will bind to pLux/cI hybrid promoter, leading the third circuit to be off.
• Circuit condition with Nosema:
When Nosema ceranae infects the bee, bee’s immune system will be activated, leading to the concentration of ROS (reactive oxygen species) to be high. In response, Bee. coli will enhance the production of oxyR, which forms a complex with ROS and binds to transcription binding site ahead of AhpCp (an oxyR-activated promoter; namely, the sensor), leading to the first circuit to be on.
Since the first circuit is on, the downstream ptet gene will be produced and bind to ptet, leading to the second circuit to be off.
After the second circuit is closed, no cI is produced, leading to the third circuit to be on (no repressor at all). And then, kill protein will be produced and kill Nosema Ceranae.
• Positive feedback:
Besides the LuxI and LuxR(which then form a complex called luxR/AHL) produced by first circuit, the third circuit will also produce LuxI and LuxR, which acts as a positive feedback and strongly enhances the production of killer protein.
The picture shows how LuxI will transfer to AHL, which then bind to LuxR to form a dimer:
• Circuit condition when Nosema is killed:
After Nosema is killed, the sensor will be off and no ptet will be produced.
Without ptet, the ptet will not be repressed, and the second circuit will be on, leading to the production of cI. After cI binds to pLux/cI hybrid promoter, the third circuit to be off. After that, the overall circuit will switch to the beginning stage when there is no Nosema.
Objectives
1. To know how much time it needs from sensing to killing the Nosema after infection
2. To know whether the pathway is effective or not
3. To know the range of kill protein concentration (from the minimal concentration which Is effective to kill Nosema to the maximal concentration which will not do harm to the bees)
System
It is assumed that AHL is abundant and thus the formation of AHL2LuxR complex is determined only by the concentration of LuxR; CI’s mechanism of binding to the promoter is assumed to act as hill effect form; kill protein will sustain a period of time before it is degraded naturally without any other types of decomposition.
Equation1
Kdptet = dissociation constant of CI
nptet = Hill coefficient of ptet
PoPSptet = promoter strength of ptet
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know mRNA_C_I production rate and when it can reach the level to translate the desired concentration.
Equation2
KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPSAhpCp = promoter strength of AhpCp
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know how AhpCp promoter strength (in PoPS) influences the production of ptet.
Equation3:
PoPSconstitutive= promoter strength of constitutive promoter (J23102)
N = number of plasmid in a single cell
V = volume of a cell
kdegmRNA = degrading constant of sensor promoter mRNA
The aim of the equation is to know the production rate ofmRNAoxyR, and choose the proper constitutive promoter for boosting OxyR's concentration.
Equation4:
KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPSAhpCp = promoter strength of AhpCp
KdLuxRAHL = dissociation constant ofLuxRAHL
nLuxRAHL = Hill coefficient of LuxRAHL
KdcI = dissociation constant of CI
ncI = Hill coefficient of CI
PoPSLuxcI = promoter strength of LuxcI hybrid promoter
kdegmRNA = degrading constant of sensor promoter Mrna
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know how AhpCp promoter strength (in PoPS) influences the production of LuxI.
Equation5:
KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPSAhpCp = promoter strength of AhpCp
KdLuxRAHL = dissociation constant ofLuxRAHL
nLuxRAHL = Hill coefficient of LuxRAHL
KdcI = dissociation constant of CI
ncI = Hill coefficient of CI
PoPSLuxcI = promoter strength of LuxcI hybrid promoter
kdegmRNA = degrading constant of sensor promoter Mrna
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know how AhpCp promoter strength (in PoPS) influences the production of LuxR.
Equation6
KdLuxRAHL = dissociation constant ofLuxRAHL
nLuxRAHL = Hill coefficient of LuxRAHL
KdcI = dissociation constant of CI
ncI = Hill coefficient of CI
PoPSLuxcI = promoter strength of LuxcI hybrid promoter
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know mRNA of kill protein production rate and when it can reach the level of the desired concentration.
to learn more about how we characterize pLuxCI, click here
Equation7
KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPSAhpCp = promoter strength of AhpCp
KdT7 = dissociation constant of T7
NT7 = Hill coefficient of T7
PoPST7 = promoter strength of T7 promoter
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know mRNA of T7 polymerase production rate and when it can reach the level to translate enough T7 polymerase.
Equation8:
KdT7 = dissociation constant of T7
NT7 = Hill coefficient of T7
PoPST7 = promoter strength of T7 promoter
KdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNA of enzyme PDC production rate and when it can reach the level to translate enough PDC.
Equation9:
KdT7 = dissociation constant of T7
NT7 = Hill coefficient of T7
PoPST7 = promoter strength of T7 promoter
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNA of enzyme ADH production rate and when it can reach the level to translate enough ADH.
Equation10
RBS = binding site strength
KdegCI = degrading constant of CI
The aim of the equation is to know the production rate of CI and when it can reach the concentration to repress LuxCI hybrid promoter.
Equation11
RBS = binding site strength
Kdegptet = degrading constant of ptet
The aim of the equation is to known ptet production rate and when it can reach the concentration to repress promoter ptet.
'Equation12
RBS = binding site strength
KAHL = LuxI to AHL rate constant
KdegLuxI = degrading constant of LuxI
The aim of the equation is to know the production rate of LuxI concerning the LuxIAHL reaction and LuxI degrading.
'Equation13
RBS = binding site strength
KmAHL =AHL to LuxI rate constant
KdegLuxR= degrading constant of LuxR
KonAHL = 2AHL + LuxR to (AHL)2/LuxR complex rate constant
KdegAHL = degrading constant of AHL
The aim of the equation is to know the production rate of LuxR concerning the 2AHL + LuxR to (AHL)2/LuxR reaction , AHL to LuxI reaction, and LuxR, AHL degrading.
'Equation14:
KAHL = LuxI to AHL rate constant
KonAHL = 2AHL + LuxR to (AHL)^2/LuxR complex rate constant
KoffAHL = (AHL)2/LuxR complex to 2AHL + LuxR rate constant
KdegAHL = degrading constant of AHL
The aim of the equation is to know the production rate of AHL concerning the LuxIAHL reaction, 2AHL + LuxR to (AHL)2/LuxR reaction, (AHL)2/LuxR complex to 2AHL + LuxR reaction and AHL degrading.
Equation17
RBS = binding site strength
PoPST7 = promoter strength of T7 promoter
N = number of plasmid in a single cell
V = volume of a cell
KdegADH = degrading constant of ADH (Acetaldehyde)
The aim of the equation is to know ADH production rate and when it can reach the concentration of ethanol pathway equilibrium.
Equation18
KAHL = LuxI to AHL rate constant
KonAHL = 2AHL + LuxR to (AHL)2/LuxR complex rate constant
KoffAHL = (AHL)2/LuxR complex to 2AHL + LuxR rate constant
KdegAHL = degrading constant of AHL
The aim of the equation is to know the production rate of AHL concerning the LuxIAHL reaction, 2AHL + LuxR to (AHL)2/LuxR reaction, (AHL)2/LuxR complex to 2AHL + LuxR reaction and AHL degrading.
Equation19
RBS = binding site strength
Kdegkill = degrading constant of kill
The aim of the equation is to knowkiller protein production rate and when it can reach the effective concentration to kill Nosema.
Results
Figure1: This picture shows kill protein concentration to time under pLux/CI hybrid promoter’s control and LuxR, LuxI’s positive feedback. The result indicates that kill protein concentration will reach its effective level and kill Nosema in time (36 hours after Nosema infection)
Figure2: This picture is a control group model. It shows that without Nosema infection (ROS concentration=0), ethanol production is low enough to be ignored.
Figure3: This is the picture of kill protein concentration to time under promoter CI’s control only(control group)without pLux/CI's regulation.The result shows that kill protein's full expression is too low to kill Nosema.
Discussion
Comparing to the ROS-level control group where ROS remains low when there’s no Nosema infection, kill protein production is highly boasted upon sensing Nosema.
The result indicates that kill protein concentration will reach its effective level and kill Nosema in time (36 hours after sensing Nosema infection, within the 5-day deadline when Nosema spores proliferates and the bee become incurable and infectious to other bees), at the given ROS changes and the enhanced OxyR expression (to learn more about our sensor model, click here).
Circuit design and improvement
Upon using pCI to negatively regulate kill protein production, results show that kill protein's full expression while no CI present is too low to kill Nosema. To solve this problem we subsidized CI promoter into pLux/CI promoter for additional positive feedback to boast kill protein’s production. As the modeling results regarding the modified circuit shown, the consequences are positive. (click here to learn more about our circuit)
Parameters:
| Model | Parameter | Description | Value | Unit | Reference |
|---|---|---|---|---|---|
| Sensor | KdegOxyR | OxyR degrading rate | 107 | M-1 x min-1 | Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol—disulfide status |
| KdOxyR* | Dissociation constant of OxyR* | 10-7.33 | M | ||
| KOxyR | OxyR producing rate constant | 5.012 x 1014 | X | ||
| nOxyR* | Hill coefficient of OxyR* | Ranging from 0.75~3.5,depends on what kind of ROS it react with | X | OxyR: A Molecular Code for Redox-Related Signaling | |
| N | Copy number | 887(cells containing 25% plasmid bearing cells) | Single piece |
| |
| Ethanol
| pyruvate | Initial concentration of pyruvate in MG1655 | 1.18 x 2 | g/L | Expression of pyruvate carboxylase enhances succinate production in Escherichia coli without affecting glucose uptake |
| nPDC | Hill coefficient of PDC | 2.1 | - | Purification, characterization and cDNA sequencing of pyruvate decarboxylase Zygosaccharomyces biporus | |
| nADH | Hill coefficient of ADH | at high pH values, 30◦C, n=1; at low temperature, n=3 | - | Evidence for co-operativity in coenzyme binding to tetrameric Sulfolobus
solfataricus alcohol dehydrogenase and its structural basis: fluorescence, kinetic and structural studies of the wild-type enzyme and non-co-operative N249Y mutant | |
| SEIR(exponential)
| b | Infection rate constant of Nosema ceranae to the suspected | 24/75 | Period(days)-1 | |
| r1 | Infection rate constant of K12 to the suspected | 3/20 | Period(days)-1 | 1. Environment protection administration executive yuan of R.O.C Medical bacteriology of J.A.T | |
| r2 | Infection rate constant of K12 to the latent | 3/20 | Period(days)-1 | 2. Environment protection administration executive yuan of R.O.C. 3. Medical bacteriology of J.A.T | |
| e | rate of the latent turns infectious | 1/4 | Period(days)-1 | ||
| u | Death rate of the infected | 1/8 | Period(days)-1 | ||
| k | Rate of intaking capsule | 24/11 | Period(days)-1 | ||
| SEIR(exponential&linear) | S | x(1) | Amount of total population | ||
| E | x(2) | Amount of suspected individuals | |||
| I | x(3) | Amount of individuals in the latent period | |||
| R | x(4) | Amount of infected individuals |
 "
"