Team:Exeter/Theory
From 2013.igem.org
Fentwistle (Talk | contribs) |
AdamThomas (Talk | contribs) |
||
| (94 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div class="row"> | <div class="row"> | ||
<div class="span" style="text-align:justify"> | <div class="span" style="text-align:justify"> | ||
| - | |||
| - | + | ==Theory== | |
| + | === Abstract === __NOTOC__ | ||
| - | + | [[File:Paint_by_coli.png|right|350px|]]Synthetic biology has led to the production of novel biosystems that are functionally beneficial. Many bacterial systems currently rely on external stimuli to induce transcription. Such induced transcription protocols often require constant monitoring of applied chemical concentrations, for example if a strain uses a [http://www.sigmaaldrich.com/life-science/molecular-biology/cloning-and-expression/vector-systems/t7-promoter-system.html T7 promoter system] that is controlled by LacI and a [http://lucigen.com/store/Expresso-Rhamnose-Cloning-and-Protein-Expression-System rhamnose expression vector], IPTG and rhamnose (both costly chemicals) will need to be monitored simultaneously. Additionally interactions and toxicity will need to be known and accounted for. Such complications lead to such chemical controls becoming undesirable for more complex systems. A triplet of NOT gated photoreceptors in <i>Escherichia coli</i>, will be used to create a system which is finely controlled using only light. This will be showcased using magenta, cyan and yellow pigments as outputs. Varying the intensity and wavelength of light projected onto <i>E. coli</i> will control the shade and colour produced, respectively. This will show the versatility of the optical control by creating a full colour bio-camera. Additionally, using bacteria to produce an image vastly increases the resolution when compared to conventional cameras, due to the micrometre scale of bacteria. | |
| - | + | === Project Description === | |
| - | + | Our project aims to produce a group of BioBricks organised into three modules; one each for red, green and blue ([http://en.wikipedia.org/wiki/Additive_color RGB]) light. To demonstrate the system we will control the production of cyan, magenta and yellow pigments, but we will also be inputting a system where future teams could have any gene as their "output" gene. We are using the CMY pigments, however RGB light could potentially be used to control the transcription of anything. | |
| - | + | ||
| - | + | The ultimate aim of our project is to use ([http://en.wikipedia.org/wiki/Subtractive_color CMY]) pigments to produce the world's first colour bio-photograph or | |
| + | [http://parts.igem.org/Coliroid coliroid]. To achieve this we will be building upon the work done by previous iGEM teams, most notably Texas/Austin 2004, Edinburgh 2010 and Uppsala 2011. | ||
If you would like to see any of our initial brainstorming - please feel free to follow this link ([https://2013.igem.org/Team:Exeter/Brainstorming Brainstorming]) | If you would like to see any of our initial brainstorming - please feel free to follow this link ([https://2013.igem.org/Team:Exeter/Brainstorming Brainstorming]) | ||
| - | == The Camera == | + | === The Camera === |
| - | Bio- | + | Bio-photography is the application of genetically engineered bacteria to act as the light sensor of a camera, akin to current [[https://en.wikipedia.org/wiki/Image_sensor CCD/CMOS]] digital sensors and photographic film. One huge advantage of using bacteria is their micrometer scale surface area. This gives them the property of being much smaller than a digital sensor, giving bio-cameras the potential to produce images with far greater resolution and pixel density than current digital photography offers. |
| - | We aim to produce the world's first colour bio-photograph by producing | + | We aim to produce the world's first colour bio-photograph by producing cyan, magenta and yellow pigments in response to red, green and blue light. We plan to combine this system with a lens to focus the image onto plated bacteria to form the world's first colour bio-camera. |
=== Colour system === | === Colour system === | ||
| - | [[File:Colour_wheels.png| | + | [[File:Colour_wheels.png|left||330px|Image: 330 pixels|Colour wheel.]] |
To produce the full colour spectrum requires all three [http://en.wikipedia.org/wiki/Primary_color primary colours]. This is because any colour can be produced by mixing the primary colours but primary colours cannot be produced by mixing any colours. | To produce the full colour spectrum requires all three [http://en.wikipedia.org/wiki/Primary_color primary colours]. This is because any colour can be produced by mixing the primary colours but primary colours cannot be produced by mixing any colours. | ||
| - | In our system we are using two sets of primary colours. | + | In our system we are using two sets of primary colours. red, green and blue for light and cyan, magenta and yellow for pigments. This is because light and pigment mix differently; light is additive and pigment is subtractive: |
Light and pigment produce colour in different ways. While coloured light has a peak wavelength corresponding to its colour, coloured pigment absorbs all wavelengths that do not correspond to its colour and so reflect the corresponding coloured light: | Light and pigment produce colour in different ways. While coloured light has a peak wavelength corresponding to its colour, coloured pigment absorbs all wavelengths that do not correspond to its colour and so reflect the corresponding coloured light: | ||
| - | + | Using [http://en.wikipedia.org/wiki/Boolean_algebra Boolean Algebra], various different colour pairing systems can be tested. If red, blue and yellow pigments were to be used, the closest pairing of inputs would require the following logic for the expression of yellow pigment: | |
| - | + | <TABLE cellpadding="1"> | |
| + | <TR> | ||
| + | <TD> [[File:Equation1.png|left||290px]] | ||
| + | </TR> | ||
| + | </TABLE> | ||
| - | + | which simplifies to | |
| - | [[Image: | + | <TABLE cellpadding="1"> |
| + | <TR> | ||
| + | <TD> [[Image:Equation2_boolean.png|left||115px]] | ||
| + | </TR> | ||
| + | </TABLE> | ||
| + | Where Y is the yellow pigment expression, R red light input; G green light input, B blue light input. But this pigment system is unable to produce cyan and magenta accurately. | ||
| - | + | The most common pigment selection for printing is CMYK (cyan, magenta, yellow, black). The boolean expression for the yellow pigment expression in this system, | |
| + | <TABLE cellpadding="1"> | ||
| + | <TR> | ||
| + | <TD> [[Image:Equation3.png|left||450px]] | ||
| + | </TR> | ||
| + | </TABLE> | ||
| + | which simplifies to | ||
| + | <TABLE cellpadding="1"> | ||
| + | <TR> | ||
| + | <TD> [[Image:Equation4.png|left||220px]] | ||
| + | </TR> | ||
| + | </TABLE> | ||
| + | The full logic gate system required for accurate expression is seen below. As can be seen, this system requires 3 NOT gates; 2 OR gates; and 4 AND gates. This would be difficult to successfully implement in a biological system. | ||
| + | <div> [[Image:Better_Logic_gates_including_black.png|center||500px|Image: 500 pixels]] </div> | ||
| + | To simplify this system, if we remove the black pigment K, the boolean expression for yellow expression becomes | ||
| + | <TABLE cellpadding="1"> | ||
| + | <TR> | ||
| + | <TD> [[Image:Equation5.png|left||600px]] | ||
| + | </TR> | ||
| + | </TABLE> | ||
| + | which simplifies very nicely to | ||
| + | <TABLE cellpadding="1"> | ||
| + | <TR> | ||
| + | <TD> [[Image:Equation6.png|left||75px]] | ||
| + | </TR> | ||
| + | </TABLE> | ||
| + | In fact the boolean expressions for all the pigment expression in the CMY system simplify. The full logic gate system for the CMY pigment system can be seen below. | ||
| + | <div>[[Image:Better_Logic_gates_not_including_black.png|center||500px|Image: 500 pixels]]</div> | ||
| - | + | This system shows red, green, and blue light repressing the production of cyan, magenta and yellow pigment respectively, theoretically being able to produce the full spectrum of colours. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | This system shows red light | + | |
<!-- Add camera design evolution: pinhole -> lens -> full fledged --> | <!-- Add camera design evolution: pinhole -> lens -> full fledged --> | ||
| + | === The Biology === | ||
| + | <TABLE cellpadding="5" align='left'> | ||
| + | <TR> | ||
| + | <TD> [[File:Light_to_Pigment_Repression.png|left||300px|]]<center>Light to pigment repression.</center> | ||
| + | </TR> | ||
| + | </TABLE> | ||
| + | The focus of our wet work is the creation of three independent 'light to output' pathways in <i>E. coli</i>. Each pathway will control the transcription of selected genes using red, green or blue light. Incident light will prevent the transcription of the selected gene. | ||
| + | In our bio-camera the controlled genes will be those that code for the production of cyan, magenta and yellow pigments: | ||
| + | * Red light prevents the production of cyan pigment | ||
| + | * Green light prevents the production of magenta pigment | ||
| + | * Blue light prevents the production of yellow pigment | ||
| + | Each sensor prevents the production of its opposing pigment, producing the pigment which is the same colour of light that the light sensor detects. The mix of pigments will reproduce the colour of light incident on the bacteria. | ||
| + | === Red light pathway === | ||
| + | <TABLE align='right'> | ||
| + | <TR> | ||
| + | <TD> [[File:Red_Pathway.png|left||500px|]] | ||
| + | </TR> | ||
| + | </TABLE> | ||
| + | This pathway uses the well characterised Cph8 red light sensor (composing of Cph1 and Envz). When there is no red light present, this protein causes the phosphorylation of OmpR, to OmpR-P. This then diffuses away from the red light sensor, and can bind to a specialised ''ompC'' promoter sequence. This is upstream of an RBS <i>B0034</i> and the <i> K592011</i> gene which is just the coding region of the cyan pigment. This is then transcribed and cyan pigment is produced. It can be noted that there has been no terminator added to this bio brick, due to there being one present in the plasmid backbone. | ||
| + | When Cph8 is exposed to red light, the phosphorylation of OmpR ceases. The OmpR is not capable of diffusing away from the red light sensor, so does not bind to the ''ompC'' promoter before the cyan pigment gene. This means that the cyan pigment can not be generated. Cyan pigment in this system will be repressed, whereas magenta and yellow pigments will still be produced, which when mixed create a red pigment. | ||
| - | + | The red light sensor has been worked on by several iGEM teams for a variety of purposes, and features in [http://www.nature.com/nature/journal/v438/n7067/full/nature04405.html several] [http://www.sciencedirect.com/science/article/pii/S0958166911007476 academic] [http://pdfs.taborlab.rice.edu/levy_ieee_2006.pdf publications]. | |
| - | + | === Green light pathway === | |
| - | + | [[File:Green_Pathway.png|left||600px|]] | |
| + | The green light module works differently to the red and blue light modules but it still uses a light sensor (CcaS), intermediate protein (CcaR) and a specialised promoter. However, when the system is exposed to green light, CcaR is phosphorylated and CcaR-P is generated, allowing the synthesis of the magenta pigment to be up regulated instead of prevented. To make an overall green colour, we only want the yellow and cyan pigments to be transcribed, so production of the magenta pigment is problematic. | ||
| - | + | To overcome this issue, we will introduce an inverter system. CcaR-P will be produced when the bacteria are exposed to green light, but instead of binding to a promoter placed before the gene coding for magenta pigment, it will bind to a cI repressor system. The subsequent transcription of the cI repressor protein will prevent the synthesis of the magenta pigment, as it will bind to a cI promoter. Binding of cI to this promoter ceases transcription of any following genes, therefore magenta will not be produced. | |
| - | + | ||
| - | + | ||
| - | [[ | + | [http://www.pnas.org/content/105/28/9528.full.pdf Several] [http://www.ncbi.nlm.nih.gov/pubmed/20404166 academic] [http://www.anselmlevskaya.com/publications/J.%20Mol.%20Biol.%202010%20Tabor.pdf papers] were referred to in characterising our green light module. |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== Blue light pathway === | === Blue light pathway === | ||
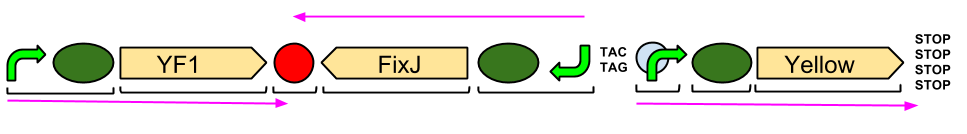
| - | The YtvA/FixL blue light sensor phosphorylates FixJ to FixJ-p in the absence of blue light. | + | [[File:Blue_Pathway.png|right||500px|]] |
| + | The YtvA/FixL blue light sensor phosphorylates FixJ to FixJ-p in the absence of blue light. FixJ-p then binds to the promoter for the yellow pigment gene and stop codon. In the absence of blue light the Yellow pigment is produced. | ||
The blue light module works in much the same way as the red light module; absence of blue light allows the phosphorylation of an intermediate protein (FixJ) by the blue light sensor (YF1). FixJ can then bind to its specialised promoter which controls the transcription of the yellow pigment (amilGFP). Exposure to blue light leads to the suppression in production of the yellow pigment. | The blue light module works in much the same way as the red light module; absence of blue light allows the phosphorylation of an intermediate protein (FixJ) by the blue light sensor (YF1). FixJ can then bind to its specialised promoter which controls the transcription of the yellow pigment (amilGFP). Exposure to blue light leads to the suppression in production of the yellow pigment. | ||
| Line 113: | Line 147: | ||
The remaining pigments, magenta and cyan, will combine to form a blue colour. | The remaining pigments, magenta and cyan, will combine to form a blue colour. | ||
| - | [[ | + | There are a [http://www.sciencedirect.com/science/article/pii/S0022283608015088 variety] of [http://onlinelibrary.wiley.com/doi/10.1002/biot.201100091/abstract;jsessionid=0D41D00C707501B442B90077720E9C82.f02t01?deniedAccessCustomisedMessage=&userIsAuthenticated=false papers] [http://www.pnas.org/content/105/31/10709.figures-only featuring] the [http://www.sciencedirect.com/science/article/pii/S1367593109001434 proteins] in our Blue Light Module. |
| + | |||
| Line 122: | Line 157: | ||
As we have the three different light modules to work with, we can experiment with different techniques and companies when it comes to our assembly techniques and synthesis. | As we have the three different light modules to work with, we can experiment with different techniques and companies when it comes to our assembly techniques and synthesis. | ||
| - | [[File: | + | [[File:Diagram of Red Plasmid for site.png|left||600px|]] |
| - | The Red Light Module is the "easiest" to work with | + | The Red Light Module is the "easiest" to work with. This is because we only need to be working with the red light sensor and cyan pigment (the intermediate protein, OmpR, is already present in the genome of <i>E. coli</i>, so we don't need to include the genes which code for it). We will try and assemble this module ourselves in the lab, using a variety of assembly techniques. The pink arrows indicate the direction of transcription. <i>ompC</i> is the specific promoter which is bound by phosphorylated OmpR, the intermediate protein. The black brackets indicate the separate BioBricks (eg. BBa_S05058 is an RBS with the gene that codes for our cyan pigment). The cyan pigment has been placed before the red light sensor so that if there is accidental read-through, we're not getting unwanted transcription of the cyan pigment. |
| - | + | [[File:Better_Diagram_of_Blue_plasmid.png|right||700px|]] | |
| - | The | + | The Blue Light Module is being synthesised for us by DNA2.0. It includes YFI (our blue light sensor), FixJ (the intermediate protein) and amilGFP (our yellow pigment). The pink arrows indicate the direction of transcription. The central section, coding for FixJ, has been flipped backwards and upside-down. This ensures that, if we get accidental read-through from the RNA polymerase from YF1, the sequence for FixJ will code for nonsense. Nothing will be made which will interfere with our system. Again, the black brackets indicate individual BioBricks. The "TACTAG" indicates a sequence of TACTAG "scars" to space out the promoter for FixJ and the promoter for the yellow pigment. This is more of a precaution than anything else. The list of "STOP" codons acts a terminator. |
| - | [[File: | + | [[File:Diagram of Green plasmid for site.png|left||700px|]] |
| - | The | + | The Green Light Module is being synthesised using IDT gBlocks. cR indicates the promoter where the cI inverter protein binds. Again, we have some TACTAG spacers between the promoters, and some sections have been flipped backwards and upside-down. The directions of transcription are shown with pink arrows. The transcripts; CcaS, CcaR, cI inverter and the magenta pigment have all been synthesised with an attached his-tag. We thought this would be useful in case we wanted to isolate and purifying any of these transcripts via a nickel column. |
| - | The | + | == The Future == |
| - | + | Our project aims to provide a foundation for the fine control of organisms using multiple wavelengths of light. The potential use of lasers to control bacteria could provide high spatial control, while a well characterised system could provide good temporal control. Importantly, response is not limited to a molecular output. Possibilities include taxis, luminescence, electricity, magnetic fields and sound. | |
| - | + | As has been proven in past research, systems expressed in ''E.coli'' can be modified and expressed in a eukaryotic model. In consideration, such a system could enable finely controlled mixed volume outputs of medically relevant molecules (e.g. patient specific antibody treatment). From an economic angle, a quantitative light input will correlate to a desired complex output from a single customizable organism strain, vastly increasing production times from current processes. Attractively, laboratories will have reduced chemical usage and only require a single processing machine, as light is a cheap, easily manipulated, and mostly harmless resource. | |
| + | High spatial and medium temporal control over each output should also be possible by the use of multi-wavelength lasers. This has many potential applications from the manufacturing of composite materials to control of engineered organisms in an environment. | ||
| - | + | As a sideline, we’re also going to test a new way of preserving and presenting bacterial culture using pouring plastics. Unfortunately, this would kill the bacteria, but should give academics a new way to physically present colonies and cultures they have been working on. For us, it will allow a way of easily transporting our “photographs” without unwieldy plates and gels. | |
| - | == | + | Considering the relatively short time frame of an iGEM project, we have had to simplify several aspects which, given more time, would have been logical to elaborate on within the bounds of our project. We would have liked to have had all three light modules (Red, Green and Blue) expressed in a single bacteria; we have had to have each module expressed in separate bacteria which are then homogeneously combined on our agar plates. As each module currently produces their corresponding pigment at different rates, we also would need to fine-tune the system to get cohesive and cooperative pigment synthesis. It would also be useful to find a red light sensor protein and intermediate protein for the Red Light Module that <i>aren't</i> involved in any other processes within <i>E. coli</i>, as our current system is also linked to osmo-sensing regulation within our competent cells. |
| + | |||
| + | ===Modelling Future=== | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
As mentioned we will be using additive and subtractive colour combinations to allow synthesis of the correct pigment output, corresponding the colours of light the bacteria are exposed to. For example the NOT gated input of red light represses synthesis of cyan pigment enabling an output of yellow and magenta pigment. We hope to analyse the absorption spectrum of each pigment, both alone and in combination. Visualisation of light sensitivity throughout the system will enable calibration of the light input ensuring reliable output. Further to this other teams reported their light sensing systems working better at temperatures less than 37 degrees, which we will also investigate. | As mentioned we will be using additive and subtractive colour combinations to allow synthesis of the correct pigment output, corresponding the colours of light the bacteria are exposed to. For example the NOT gated input of red light represses synthesis of cyan pigment enabling an output of yellow and magenta pigment. We hope to analyse the absorption spectrum of each pigment, both alone and in combination. Visualisation of light sensitivity throughout the system will enable calibration of the light input ensuring reliable output. Further to this other teams reported their light sensing systems working better at temperatures less than 37 degrees, which we will also investigate. | ||
| - | |||
| - | |||
| - | + | For the blue and red light sensors, the presence of their corresponding wavelength of light causes auto-phosphorylation of an intermediate protein. Phosphorylated intermediates then freely bind as repressors to the corresponding output gene (cyan for red light, yellow for blue light). With the green light sensor, a signal inverter will be introduced, as the FixJ system acts as an activator not a repressor (the exact opposite of what we want). Instead of acting on the gene coding for the output (magenta) the phosphorylated intermediate binds instead to a gene coding for the cI protein used in the lambda phage (a bacteriophage which infects E. coli). Once synthesised the cl protein acts as the output (magenta) repressor. | |
| - | + | ||
| - | + | If you would like to see how our project went, please feel free to visit the [https://2013.igem.org/Team:Exeter/Results Results] page of our website. | |
| - | + | ||
| - | + | ===Potential Applications=== | |
| - | + | ||
| + | |||
| + | Our project is potentially relevant and applicable to many areas within technology, industry and even medicine. One of our fundamental aims was to design a system that would allow other teams or users to use any gene they desired as the set output gene, thus allowing progression and greater freedom whilst also accommodating a range of other projects. The development of a true bacterial camera would also hold its own niche alongside other paths of photography, it even presents a number of positive advancements on standard digital photography. Incredibly high resolution images are possible, whilst noise problems could potentially be avoided and the technology may even have applications for low light photography. Light targeted therapies could be introduced that use bacteria inserted into a wound or other area of the body to produce the relevant drug or treatment. This would be very beneficial in the strive for ever less invasive methods of treatment. | ||
</div> | </div> | ||
Latest revision as of 03:37, 5 October 2013
Theory
Abstract
Synthetic biology has led to the production of novel biosystems that are functionally beneficial. Many bacterial systems currently rely on external stimuli to induce transcription. Such induced transcription protocols often require constant monitoring of applied chemical concentrations, for example if a strain uses a [http://www.sigmaaldrich.com/life-science/molecular-biology/cloning-and-expression/vector-systems/t7-promoter-system.html T7 promoter system] that is controlled by LacI and a [http://lucigen.com/store/Expresso-Rhamnose-Cloning-and-Protein-Expression-System rhamnose expression vector], IPTG and rhamnose (both costly chemicals) will need to be monitored simultaneously. Additionally interactions and toxicity will need to be known and accounted for. Such complications lead to such chemical controls becoming undesirable for more complex systems. A triplet of NOT gated photoreceptors in Escherichia coli, will be used to create a system which is finely controlled using only light. This will be showcased using magenta, cyan and yellow pigments as outputs. Varying the intensity and wavelength of light projected onto E. coli will control the shade and colour produced, respectively. This will show the versatility of the optical control by creating a full colour bio-camera. Additionally, using bacteria to produce an image vastly increases the resolution when compared to conventional cameras, due to the micrometre scale of bacteria.Project Description
Our project aims to produce a group of BioBricks organised into three modules; one each for red, green and blue ([http://en.wikipedia.org/wiki/Additive_color RGB]) light. To demonstrate the system we will control the production of cyan, magenta and yellow pigments, but we will also be inputting a system where future teams could have any gene as their "output" gene. We are using the CMY pigments, however RGB light could potentially be used to control the transcription of anything.
The ultimate aim of our project is to use ([http://en.wikipedia.org/wiki/Subtractive_color CMY]) pigments to produce the world's first colour bio-photograph or [http://parts.igem.org/Coliroid coliroid]. To achieve this we will be building upon the work done by previous iGEM teams, most notably Texas/Austin 2004, Edinburgh 2010 and Uppsala 2011.
If you would like to see any of our initial brainstorming - please feel free to follow this link (Brainstorming)
The Camera
Bio-photography is the application of genetically engineered bacteria to act as the light sensor of a camera, akin to current [CCD/CMOS] digital sensors and photographic film. One huge advantage of using bacteria is their micrometer scale surface area. This gives them the property of being much smaller than a digital sensor, giving bio-cameras the potential to produce images with far greater resolution and pixel density than current digital photography offers.
We aim to produce the world's first colour bio-photograph by producing cyan, magenta and yellow pigments in response to red, green and blue light. We plan to combine this system with a lens to focus the image onto plated bacteria to form the world's first colour bio-camera.
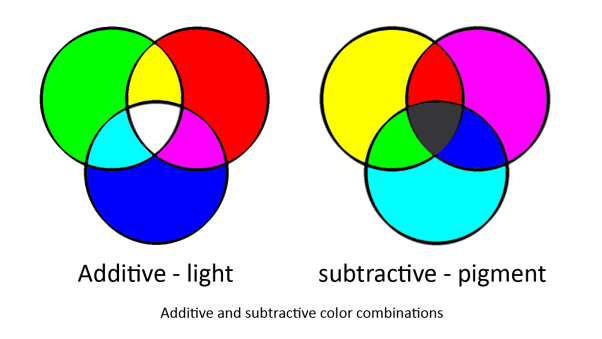
Colour system
To produce the full colour spectrum requires all three [http://en.wikipedia.org/wiki/Primary_color primary colours]. This is because any colour can be produced by mixing the primary colours but primary colours cannot be produced by mixing any colours.
In our system we are using two sets of primary colours. red, green and blue for light and cyan, magenta and yellow for pigments. This is because light and pigment mix differently; light is additive and pigment is subtractive:
Light and pigment produce colour in different ways. While coloured light has a peak wavelength corresponding to its colour, coloured pigment absorbs all wavelengths that do not correspond to its colour and so reflect the corresponding coloured light:
Using [http://en.wikipedia.org/wiki/Boolean_algebra Boolean Algebra], various different colour pairing systems can be tested. If red, blue and yellow pigments were to be used, the closest pairing of inputs would require the following logic for the expression of yellow pigment:
which simplifies to
Where Y is the yellow pigment expression, R red light input; G green light input, B blue light input. But this pigment system is unable to produce cyan and magenta accurately.
The most common pigment selection for printing is CMYK (cyan, magenta, yellow, black). The boolean expression for the yellow pigment expression in this system,
which simplifies to
The full logic gate system required for accurate expression is seen below. As can be seen, this system requires 3 NOT gates; 2 OR gates; and 4 AND gates. This would be difficult to successfully implement in a biological system.
To simplify this system, if we remove the black pigment K, the boolean expression for yellow expression becomes
which simplifies very nicely to
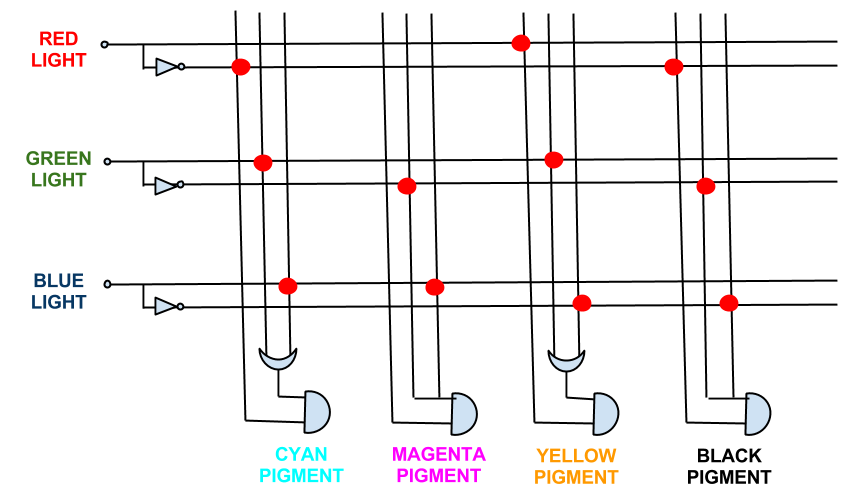
In fact the boolean expressions for all the pigment expression in the CMY system simplify. The full logic gate system for the CMY pigment system can be seen below.
This system shows red, green, and blue light repressing the production of cyan, magenta and yellow pigment respectively, theoretically being able to produce the full spectrum of colours.
The Biology
| |
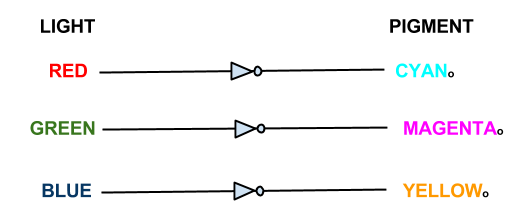
The focus of our wet work is the creation of three independent 'light to output' pathways in E. coli. Each pathway will control the transcription of selected genes using red, green or blue light. Incident light will prevent the transcription of the selected gene.
In our bio-camera the controlled genes will be those that code for the production of cyan, magenta and yellow pigments:
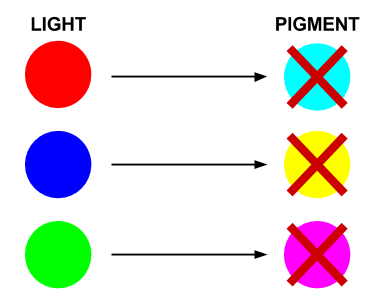
- Red light prevents the production of cyan pigment
- Green light prevents the production of magenta pigment
- Blue light prevents the production of yellow pigment
Each sensor prevents the production of its opposing pigment, producing the pigment which is the same colour of light that the light sensor detects. The mix of pigments will reproduce the colour of light incident on the bacteria.
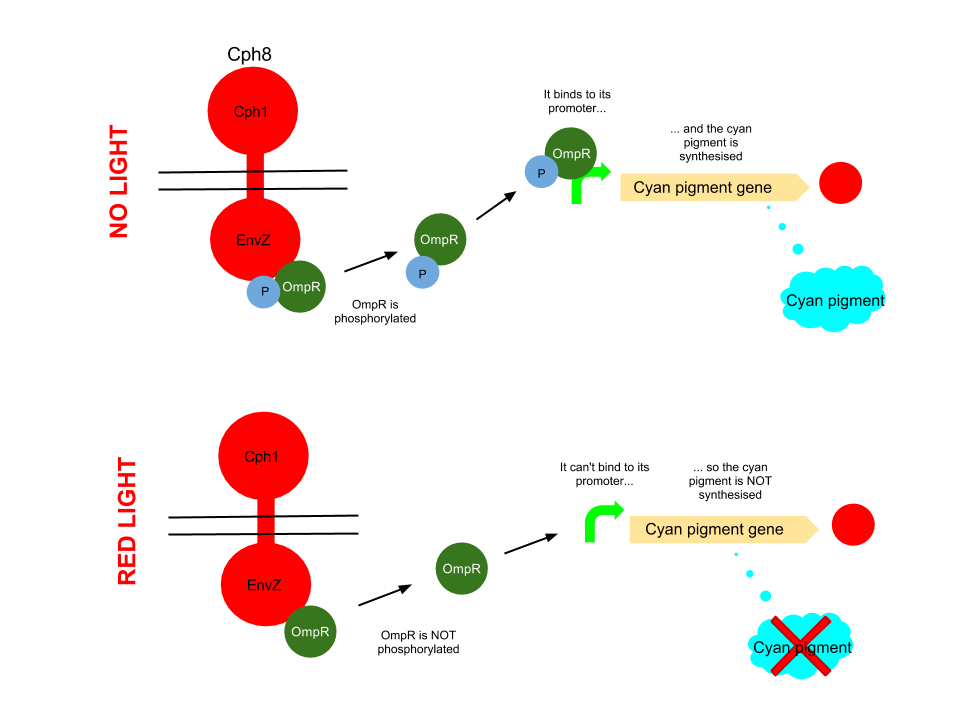
Red light pathway
This pathway uses the well characterised Cph8 red light sensor (composing of Cph1 and Envz). When there is no red light present, this protein causes the phosphorylation of OmpR, to OmpR-P. This then diffuses away from the red light sensor, and can bind to a specialised ompC promoter sequence. This is upstream of an RBS B0034 and the K592011 gene which is just the coding region of the cyan pigment. This is then transcribed and cyan pigment is produced. It can be noted that there has been no terminator added to this bio brick, due to there being one present in the plasmid backbone.
When Cph8 is exposed to red light, the phosphorylation of OmpR ceases. The OmpR is not capable of diffusing away from the red light sensor, so does not bind to the ompC promoter before the cyan pigment gene. This means that the cyan pigment can not be generated. Cyan pigment in this system will be repressed, whereas magenta and yellow pigments will still be produced, which when mixed create a red pigment.
The red light sensor has been worked on by several iGEM teams for a variety of purposes, and features in [http://www.nature.com/nature/journal/v438/n7067/full/nature04405.html several] [http://www.sciencedirect.com/science/article/pii/S0958166911007476 academic] [http://pdfs.taborlab.rice.edu/levy_ieee_2006.pdf publications].
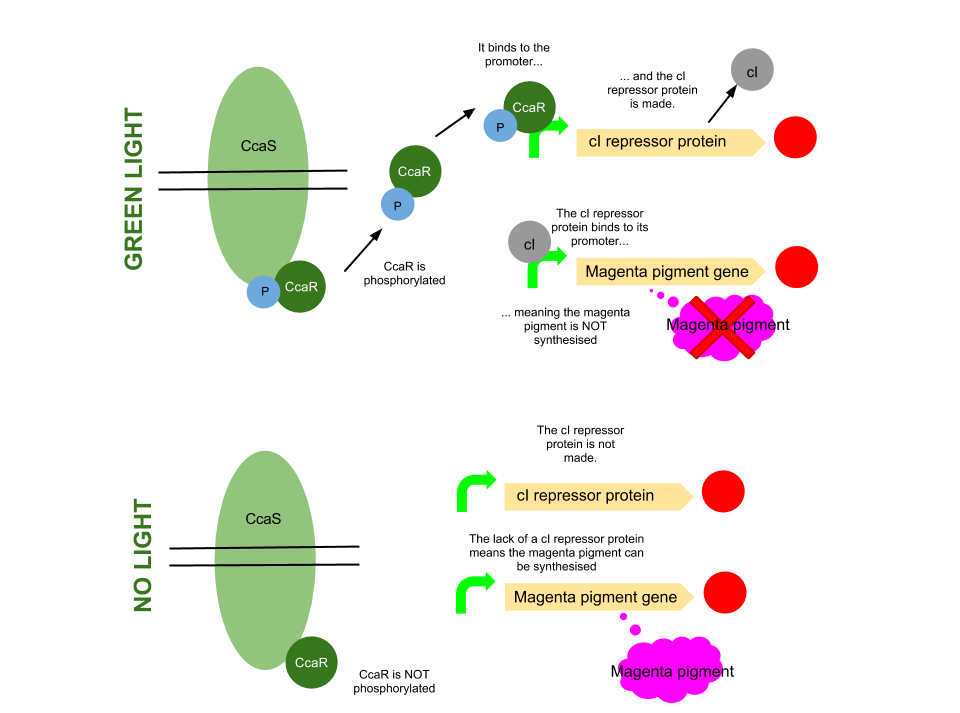
Green light pathway
The green light module works differently to the red and blue light modules but it still uses a light sensor (CcaS), intermediate protein (CcaR) and a specialised promoter. However, when the system is exposed to green light, CcaR is phosphorylated and CcaR-P is generated, allowing the synthesis of the magenta pigment to be up regulated instead of prevented. To make an overall green colour, we only want the yellow and cyan pigments to be transcribed, so production of the magenta pigment is problematic.
To overcome this issue, we will introduce an inverter system. CcaR-P will be produced when the bacteria are exposed to green light, but instead of binding to a promoter placed before the gene coding for magenta pigment, it will bind to a cI repressor system. The subsequent transcription of the cI repressor protein will prevent the synthesis of the magenta pigment, as it will bind to a cI promoter. Binding of cI to this promoter ceases transcription of any following genes, therefore magenta will not be produced.
[http://www.pnas.org/content/105/28/9528.full.pdf Several] [http://www.ncbi.nlm.nih.gov/pubmed/20404166 academic] [http://www.anselmlevskaya.com/publications/J.%20Mol.%20Biol.%202010%20Tabor.pdf papers] were referred to in characterising our green light module.
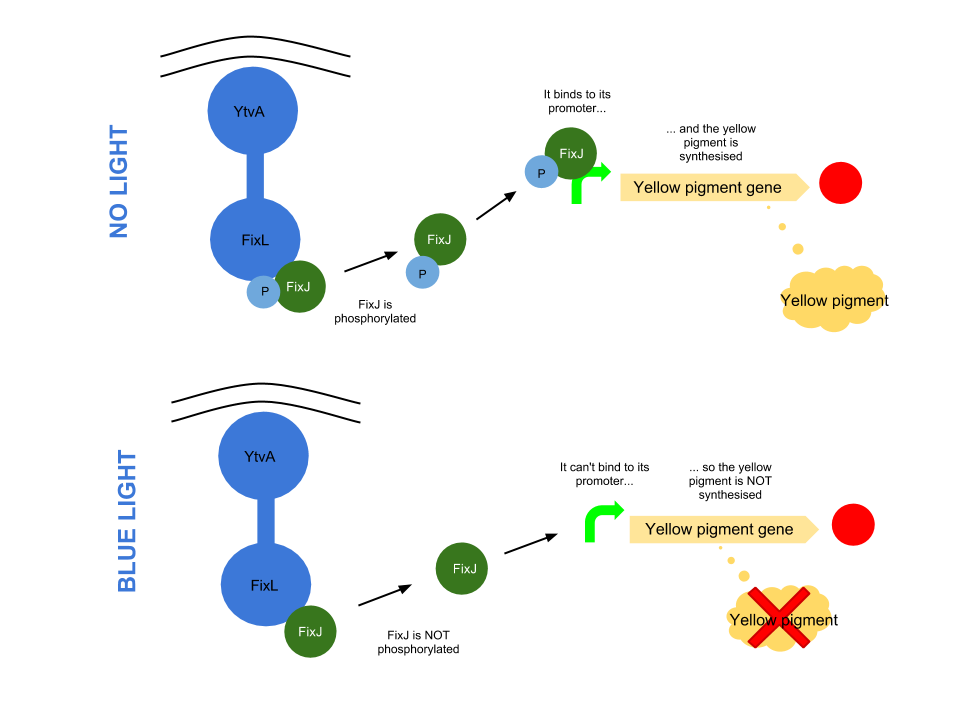
Blue light pathway
The YtvA/FixL blue light sensor phosphorylates FixJ to FixJ-p in the absence of blue light. FixJ-p then binds to the promoter for the yellow pigment gene and stop codon. In the absence of blue light the Yellow pigment is produced.
The blue light module works in much the same way as the red light module; absence of blue light allows the phosphorylation of an intermediate protein (FixJ) by the blue light sensor (YF1). FixJ can then bind to its specialised promoter which controls the transcription of the yellow pigment (amilGFP). Exposure to blue light leads to the suppression in production of the yellow pigment.
The remaining pigments, magenta and cyan, will combine to form a blue colour.
There are a [http://www.sciencedirect.com/science/article/pii/S0022283608015088 variety] of [http://onlinelibrary.wiley.com/doi/10.1002/biot.201100091/abstract;jsessionid=0D41D00C707501B442B90077720E9C82.f02t01?deniedAccessCustomisedMessage=&userIsAuthenticated=false papers] [http://www.pnas.org/content/105/31/10709.figures-only featuring] the [http://www.sciencedirect.com/science/article/pii/S1367593109001434 proteins] in our Blue Light Module.
Our constructs
As we have the three different light modules to work with, we can experiment with different techniques and companies when it comes to our assembly techniques and synthesis.
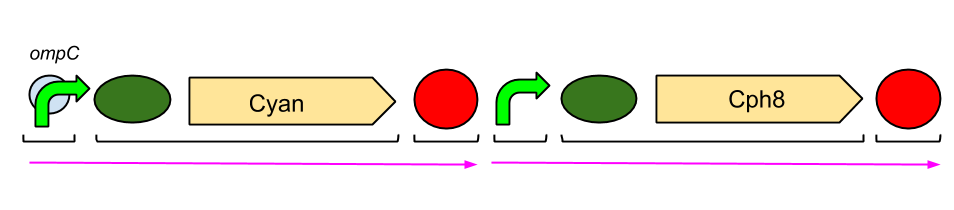
The Red Light Module is the "easiest" to work with. This is because we only need to be working with the red light sensor and cyan pigment (the intermediate protein, OmpR, is already present in the genome of E. coli, so we don't need to include the genes which code for it). We will try and assemble this module ourselves in the lab, using a variety of assembly techniques. The pink arrows indicate the direction of transcription. ompC is the specific promoter which is bound by phosphorylated OmpR, the intermediate protein. The black brackets indicate the separate BioBricks (eg. BBa_S05058 is an RBS with the gene that codes for our cyan pigment). The cyan pigment has been placed before the red light sensor so that if there is accidental read-through, we're not getting unwanted transcription of the cyan pigment.
The Blue Light Module is being synthesised for us by DNA2.0. It includes YFI (our blue light sensor), FixJ (the intermediate protein) and amilGFP (our yellow pigment). The pink arrows indicate the direction of transcription. The central section, coding for FixJ, has been flipped backwards and upside-down. This ensures that, if we get accidental read-through from the RNA polymerase from YF1, the sequence for FixJ will code for nonsense. Nothing will be made which will interfere with our system. Again, the black brackets indicate individual BioBricks. The "TACTAG" indicates a sequence of TACTAG "scars" to space out the promoter for FixJ and the promoter for the yellow pigment. This is more of a precaution than anything else. The list of "STOP" codons acts a terminator.
The Green Light Module is being synthesised using IDT gBlocks. cR indicates the promoter where the cI inverter protein binds. Again, we have some TACTAG spacers between the promoters, and some sections have been flipped backwards and upside-down. The directions of transcription are shown with pink arrows. The transcripts; CcaS, CcaR, cI inverter and the magenta pigment have all been synthesised with an attached his-tag. We thought this would be useful in case we wanted to isolate and purifying any of these transcripts via a nickel column.
The Future
Our project aims to provide a foundation for the fine control of organisms using multiple wavelengths of light. The potential use of lasers to control bacteria could provide high spatial control, while a well characterised system could provide good temporal control. Importantly, response is not limited to a molecular output. Possibilities include taxis, luminescence, electricity, magnetic fields and sound.
As has been proven in past research, systems expressed in E.coli can be modified and expressed in a eukaryotic model. In consideration, such a system could enable finely controlled mixed volume outputs of medically relevant molecules (e.g. patient specific antibody treatment). From an economic angle, a quantitative light input will correlate to a desired complex output from a single customizable organism strain, vastly increasing production times from current processes. Attractively, laboratories will have reduced chemical usage and only require a single processing machine, as light is a cheap, easily manipulated, and mostly harmless resource. High spatial and medium temporal control over each output should also be possible by the use of multi-wavelength lasers. This has many potential applications from the manufacturing of composite materials to control of engineered organisms in an environment.
As a sideline, we’re also going to test a new way of preserving and presenting bacterial culture using pouring plastics. Unfortunately, this would kill the bacteria, but should give academics a new way to physically present colonies and cultures they have been working on. For us, it will allow a way of easily transporting our “photographs” without unwieldy plates and gels.
Considering the relatively short time frame of an iGEM project, we have had to simplify several aspects which, given more time, would have been logical to elaborate on within the bounds of our project. We would have liked to have had all three light modules (Red, Green and Blue) expressed in a single bacteria; we have had to have each module expressed in separate bacteria which are then homogeneously combined on our agar plates. As each module currently produces their corresponding pigment at different rates, we also would need to fine-tune the system to get cohesive and cooperative pigment synthesis. It would also be useful to find a red light sensor protein and intermediate protein for the Red Light Module that aren't involved in any other processes within E. coli, as our current system is also linked to osmo-sensing regulation within our competent cells.
Modelling Future
As mentioned we will be using additive and subtractive colour combinations to allow synthesis of the correct pigment output, corresponding the colours of light the bacteria are exposed to. For example the NOT gated input of red light represses synthesis of cyan pigment enabling an output of yellow and magenta pigment. We hope to analyse the absorption spectrum of each pigment, both alone and in combination. Visualisation of light sensitivity throughout the system will enable calibration of the light input ensuring reliable output. Further to this other teams reported their light sensing systems working better at temperatures less than 37 degrees, which we will also investigate.
For the blue and red light sensors, the presence of their corresponding wavelength of light causes auto-phosphorylation of an intermediate protein. Phosphorylated intermediates then freely bind as repressors to the corresponding output gene (cyan for red light, yellow for blue light). With the green light sensor, a signal inverter will be introduced, as the FixJ system acts as an activator not a repressor (the exact opposite of what we want). Instead of acting on the gene coding for the output (magenta) the phosphorylated intermediate binds instead to a gene coding for the cI protein used in the lambda phage (a bacteriophage which infects E. coli). Once synthesised the cl protein acts as the output (magenta) repressor.
If you would like to see how our project went, please feel free to visit the Results page of our website.
Potential Applications
Our project is potentially relevant and applicable to many areas within technology, industry and even medicine. One of our fundamental aims was to design a system that would allow other teams or users to use any gene they desired as the set output gene, thus allowing progression and greater freedom whilst also accommodating a range of other projects. The development of a true bacterial camera would also hold its own niche alongside other paths of photography, it even presents a number of positive advancements on standard digital photography. Incredibly high resolution images are possible, whilst noise problems could potentially be avoided and the technology may even have applications for low light photography. Light targeted therapies could be introduced that use bacteria inserted into a wound or other area of the body to produce the relevant drug or treatment. This would be very beneficial in the strive for ever less invasive methods of treatment.
 "
"