Team:Uppsala/safety-experiment
From 2013.igem.org
(Difference between revisions)
| (13 intermediate revisions not shown) | |||
| Line 17: | Line 17: | ||
/*Add favicon*/ $("link[rel='shortcut icon']").remove(); $("head").append("<link rel='shortcut icon' type='image/png' href='https://static.igem.org/mediawiki/2013/6/6d/Uppsalas-cow-con.png'>"); }); | /*Add favicon*/ $("link[rel='shortcut icon']").remove(); $("head").append("<link rel='shortcut icon' type='image/png' href='https://static.igem.org/mediawiki/2013/6/6d/Uppsalas-cow-con.png'>"); }); | ||
</script> | </script> | ||
| + | |||
| + | <link href="https://2013.igem.org/Team:Uppsala/lightbox-css-code.css?action=raw&ctype=text/css" type="text/css" rel="stylesheet"> | ||
| + | |||
| + | <script src="https://2013.igem.org/Team:Uppsala/jquery-code.js?action=raw&ctype=text/javascript" type="text/javascript"></script> | ||
| + | |||
| + | <script src="https://2013.igem.org/Team:Uppsala/lightbox-code.js?action=raw&ctype=text/javascript" type="text/javascript"></script> | ||
</head> | </head> | ||
| Line 33: | Line 39: | ||
<div id="igem"><a href="https://2013.igem.org/Main_Page"><img class="igem" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_Uppsala_IGEM_log_blue.png"></a></div> | <div id="igem"><a href="https://2013.igem.org/Main_Page"><img class="igem" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_Uppsala_IGEM_log_blue.png"></a></div> | ||
| + | |||
| + | <img class="slogan" src="https://static.igem.org/mediawiki/2013/a/ad/Uppsala2013_Header.png"> | ||
</div> | </div> | ||
| Line 60: | Line 68: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/metabolic-engineering">Metabolic engineering</a> | <li><a href="https://2013.igem.org/Team:Uppsala/metabolic-engineering">Metabolic engineering</a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/p-coumaric-acid"> | + | <li><a href="https://2013.igem.org/Team:Uppsala/p-coumaric-acid">p-Coumaric acid</a></li> |
<li><a href="https://2013.igem.org/Team:Uppsala/resveratrol">Resveratrol</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/resveratrol">Resveratrol</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/lycopene">Lycopene</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/lycopene">Lycopene</a></li> | ||
| Line 81: | Line 89: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/modeling" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/6/63/Uppsala2013_Modeling.png"></a> | <li><a href="https://2013.igem.org/Team:Uppsala/modeling" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/6/63/Uppsala2013_Modeling.png"></a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/P-Coumaric-acid-pathway"> | + | <li><a href="https://2013.igem.org/Team:Uppsala/P-Coumaric-acid-pathway">Kinetic model</a></li> |
<li><a href="https://2013.igem.org/Team:Uppsala/modeling-tutorial">Modeling tutorial </a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/modeling-tutorial">Modeling tutorial </a></li> | ||
| + | |||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/toxicity-model">Toxicity model</a></li> | ||
</ul></li> | </ul></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/parts" id="list_type2"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/e/eb/Uppsala2013_parts.png"></a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/parts" id="list_type2"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/e/eb/Uppsala2013_parts.png"></a></li> | ||
| Line 91: | Line 101: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/carotenoid-group">Carotenoid group</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/carotenoid-group">Carotenoid group</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/chassi-group">Chassi group</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/chassi-group">Chassi group</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/advisors">Advisors</a></li> | ||
</ul></li> | </ul></li> | ||
| Line 100: | Line 111: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/public-opinion">Public opinion </a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/public-opinion">Public opinion </a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/Outreach">High school & media </a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/Outreach">High school & media </a></li> | ||
| - | + | <li><a href="https://2013.igem.org/Team:Uppsala/bioart">BioArt</a></li> | |
| + | <li><a href="https://2013.igem.org/Team:Uppsala/LactonutritiousWorld">A LactoWorld</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/killswitches">Killswitches</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/realization">Patent</a></li> | ||
</ul></li> | </ul></li> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a | + | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a></li> |
| - | + | ||
| - | + | ||
| - | + | ||
<li><a href="https://2013.igem.org/Team:Uppsala/notebook" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/3/36/Uppsala2013_Notebook.png"></a> | <li><a href="https://2013.igem.org/Team:Uppsala/notebook" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/3/36/Uppsala2013_Notebook.png"></a> | ||
<ul> | <ul> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/safety-form">Safety form</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/safety-form">Safety form</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/protocols">Protocols</a></li> | ||
</ul></li> | </ul></li> | ||
</ul> | </ul> | ||
| Line 115: | Line 127: | ||
</div> | </div> | ||
<div id="main_content"> | <div id="main_content"> | ||
| - | <h1 class="main-title"> | + | <h1 class="main-title">Safety experiment</h1> |
| + | |||
<h1>Can our modified probiotic bacteria survive and spread in the ecosystem?</h1> | <h1>Can our modified probiotic bacteria survive and spread in the ecosystem?</h1> | ||
In our human practice project we discussed possible biosafety risks that could come with the use of a genetically engineered probiotic bacteria. One of the strongest arguments against genetically engineered organisms is the risk of spreading. When incorporating GMO in our daily food, the risk of our bacteria coming in contact with our ecosystem is high. What would happen if our bacteria leaked in nature? | In our human practice project we discussed possible biosafety risks that could come with the use of a genetically engineered probiotic bacteria. One of the strongest arguments against genetically engineered organisms is the risk of spreading. When incorporating GMO in our daily food, the risk of our bacteria coming in contact with our ecosystem is high. What would happen if our bacteria leaked in nature? | ||
| - | In this simple test, we took our biobrick tyrosine ammonia lyase, <a href="http://parts.igem.org/Part:BBa_K1033000">BBa_K1033000</a>, and expressed it in our probiotic | + | In this simple test, we took our biobrick tyrosine ammonia lyase, <a href="http://parts.igem.org/Part:BBa_K1033000">BBa_K1033000</a>, and expressed it in our probiotic E. coli nissle. We wanted to see if it would be able to compete in an environment together with the wild type of the same strain. We also did an experiment to see whether the engineered strain would lose the biobrick plasmid without competition over time. |
| + | <br><br> | ||
| + | |||
<h1>The experiment</h1> | <h1>The experiment</h1> | ||
| - | We grew our nissle containing the TAL biobrick together with wild type nissle (mixed 1:1) in antiobiotic free LB medium. The culture was maintaned by a thousandfold dilution in new medium every day, and letting it grow again overnight. To be able to track the fraction of each population over time, a dilution of the culture was plated on both selective, and non selective plates. The amount of colonies on the non selective plates gives us the total amount of cells, while the amount of colonies on the selective plates gives us the number of cells carrying the biobrick plasmid. The fraction of cells carrying the plasmid declined rapidly, and already after 5 days (approximately 50 generations of growth), we saw no trace of our | + | We grew our nissle containing the TAL biobrick together with wild type nissle (mixed 1:1) in antiobiotic free LB medium. The culture was maintaned by a thousandfold dilution in new medium every day, and letting it grow again overnight. To be able to track the fraction of each population over time, a dilution of the culture was plated on both selective, and non selective plates. The amount of colonies on the non selective plates gives us the total amount of cells, while the amount of colonies on the selective plates gives us the number of cells carrying the biobrick plasmid. The fraction of cells carrying the plasmid declined rapidly, and already after 5 days (approximately 50 generations of growth), we saw no trace of our E. coli nissle with TAL on the selective plates, while the wild type still grew on the non selective plates. This clearly shows that our genetically engineered probiotic have a hard time competing against the wild type, and if released into the environment, they would most likely be outcompeted by other bacteria within days. |
| - | + | <br><br> | |
| - | + | ||
| - | <div class="safety_box"><img class="safety_pic" src="https://static.igem.org/mediawiki/2013/ | + | <div class="safety_box"><a href="https://static.igem.org/mediawiki/2013/4/4b/Uppsala2013_Safety.png" data-lightbox="roadtrip" title="Picture of our plates with E. coli nissle and E. coli nissle containing our biobrick, tyrosine ammonia lyase on a resistance plasmid. Gradually our biobrick contaning E. coli is outcompeted by the wild type E. coli nissle. To the right is an antiobiotic free plate with a positive control from one of the last days showing that the E. coli nissle without resistance plasmid is still healthy and growing."><img class="safety_pic" src="https://static.igem.org/mediawiki/2013/4/4b/Uppsala2013_Safety.png"> |
| + | <a href="https://static.igem.org/mediawiki/2013/4/49/Prime_bactus_uppsala_st.jpg" data-lightbox="roadtrip" title="Picture of our plates with E. coli nissle and E. coli nissle containing our biobrick, tyrosine ammonia lyase on a resistance plasmid. Gradually our biobrick contaning E. coli is outcompeted by the wild type E. coli nissle. To the right is an antiobiotic free plate with a positive control from one of the last days showing that the E. coli nissle without resistance plasmid is still healthy and growing."><img class="safety_pic" src="https://static.igem.org/mediawiki/2013/4/49/Prime_bactus_uppsala_st.jpg"></a></div> | ||
| - | <b><i>Figure 1.</b>Picture of our plates with | + | <b><i>Figure 1.</b> Picture of our plates with E. coli nissle and E. coli nissle containing our biobrick, tyrosine ammonia lyase on a resistance plasmid. Gradually our biobrick contaning E. coli is outcompeted by the wild type E. coli nissle. To the right is an antiobiotic free plate with a positive control from one of the last days showing that the E. coli nissle without resistance plasmid is still healthy and growing. </i> |
<br><br><br> | <br><br><br> | ||
<div id="safety_table"> | <div id="safety_table"> | ||
<b>Competition experiments between TAL and wild type</b><br> | <b>Competition experiments between TAL and wild type</b><br> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/6/6f/Table_safety_uppsala.png"><br> | + | <a href="https://static.igem.org/mediawiki/2013/6/6f/Table_safety_uppsala.png" data-lightbox="roadtrip" title="Competition experiments between E. coli Nissle carrying the TAL plasmid and wild type Nissle"><img src="https://static.igem.org/mediawiki/2013/6/6f/Table_safety_uppsala.png"></a><br> |
| - | <b><i>Figure 2.</b>Competition experiments between E. coli Nissle carrying the TAL plasmid and wild type Nissle</i> | + | <b><i>Figure 2.</b> Competition experiments between E. coli Nissle carrying the TAL plasmid and wild type Nissle</i> |
| - | <div | + | </div><br><br> |
| - | In our second experiment, when growing our bacteria | + | In our second experiment, when growing our bacteria E. coli nissle for several days in antibiotic free medium, we could not observe any loss of plasmid. This means that the plasmid is stable in Nissle, and that the sensistive bacteria in the competition experiment are true wild type bacteria, and not bacteria that initally carried the TAL plasmid but lost it during the course of the competition.<br><br> |
By plotting the competition data, with the ratio TAL/wt on the y axis and the time (measured as generations of growth) on the x axis, it is possible to determine the difference in growth rate between the strains. Our data shows that the wild type grows approximately 7.7% faster than the same strain carrying the TAL plasmid. | By plotting the competition data, with the ratio TAL/wt on the y axis and the time (measured as generations of growth) on the x axis, it is possible to determine the difference in growth rate between the strains. Our data shows that the wild type grows approximately 7.7% faster than the same strain carrying the TAL plasmid. | ||
<br><br> | <br><br> | ||
| Line 146: | Line 161: | ||
<div id="bottom-pic"> | <div id="bottom-pic"> | ||
| - | + | ||
</div> | </div> | ||
Latest revision as of 16:02, 28 October 2013
Safety experiment
Can our modified probiotic bacteria survive and spread in the ecosystem?
In our human practice project we discussed possible biosafety risks that could come with the use of a genetically engineered probiotic bacteria. One of the strongest arguments against genetically engineered organisms is the risk of spreading. When incorporating GMO in our daily food, the risk of our bacteria coming in contact with our ecosystem is high. What would happen if our bacteria leaked in nature? In this simple test, we took our biobrick tyrosine ammonia lyase, BBa_K1033000, and expressed it in our probiotic E. coli nissle. We wanted to see if it would be able to compete in an environment together with the wild type of the same strain. We also did an experiment to see whether the engineered strain would lose the biobrick plasmid without competition over time.The experiment
We grew our nissle containing the TAL biobrick together with wild type nissle (mixed 1:1) in antiobiotic free LB medium. The culture was maintaned by a thousandfold dilution in new medium every day, and letting it grow again overnight. To be able to track the fraction of each population over time, a dilution of the culture was plated on both selective, and non selective plates. The amount of colonies on the non selective plates gives us the total amount of cells, while the amount of colonies on the selective plates gives us the number of cells carrying the biobrick plasmid. The fraction of cells carrying the plasmid declined rapidly, and already after 5 days (approximately 50 generations of growth), we saw no trace of our E. coli nissle with TAL on the selective plates, while the wild type still grew on the non selective plates. This clearly shows that our genetically engineered probiotic have a hard time competing against the wild type, and if released into the environment, they would most likely be outcompeted by other bacteria within days.Figure 1. Picture of our plates with E. coli nissle and E. coli nissle containing our biobrick, tyrosine ammonia lyase on a resistance plasmid. Gradually our biobrick contaning E. coli is outcompeted by the wild type E. coli nissle. To the right is an antiobiotic free plate with a positive control from one of the last days showing that the E. coli nissle without resistance plasmid is still healthy and growing.
Competition experiments between TAL and wild type
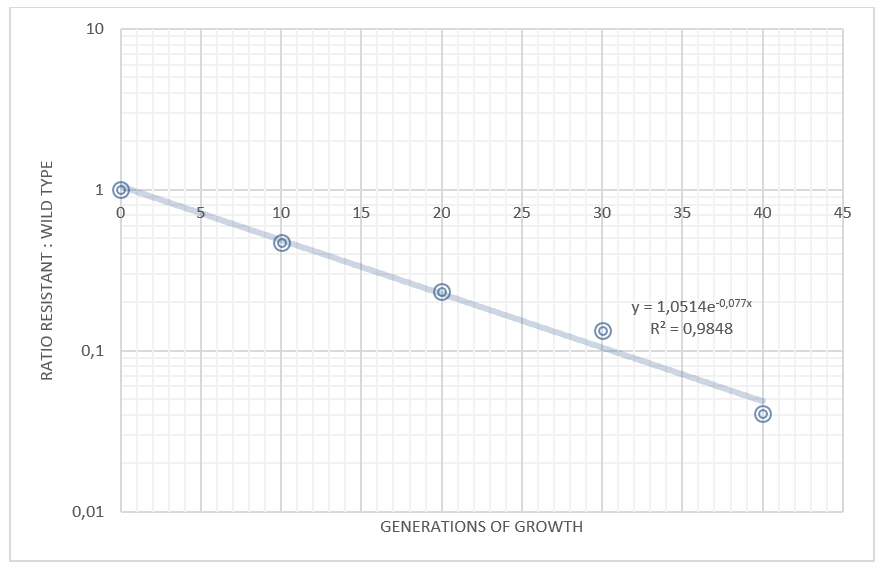
Figure 2. Competition experiments between E. coli Nissle carrying the TAL plasmid and wild type Nissle

Figure 2. Competition experiments between E. coli Nissle carrying the TAL plasmid and wild type Nissle
In our second experiment, when growing our bacteria E. coli nissle for several days in antibiotic free medium, we could not observe any loss of plasmid. This means that the plasmid is stable in Nissle, and that the sensistive bacteria in the competition experiment are true wild type bacteria, and not bacteria that initally carried the TAL plasmid but lost it during the course of the competition.
By plotting the competition data, with the ratio TAL/wt on the y axis and the time (measured as generations of growth) on the x axis, it is possible to determine the difference in growth rate between the strains. Our data shows that the wild type grows approximately 7.7% faster than the same strain carrying the TAL plasmid.
 "
"












