Team:Uppsala/miraculin
From 2013.igem.org
(Difference between revisions)
Sabrijamal (Talk | contribs) |
|||
| (23 intermediate revisions not shown) | |||
| Line 12: | Line 12: | ||
</style> | </style> | ||
| + | |||
| + | <script type="text/javascript"> | ||
| + | /*Acts once the document is loaded*/ $(document).ready(function() { /*Move the tn-main-wrap as child of <body>*/ $("#tn-main-wrap-wrap").prependTo($("body")); /*Move the menubars in the tn-main-wrap*/ $("#menubar.left-menu").appendTo($("#tn-main-wrap")); $("#menubar.right-menu").appendTo($("#tn-main-wrap")); | ||
| + | /*Spoiler JS*/ $(".tn-spoiler div").slideUp(); $(".tn-spoiler a").click(function(e) { e.preventDefault(); $(".tn-spoiler-active").removeClass("tn-spoiler-active"); $(this).parent().addClass("tn-spoiler-active"); $(".tn-spoiler").not(".tn-spoiler-active").children("div").slideUp(); $(".tn-spoiler-active").children("div").slideToggle(); }); | ||
| + | /*Add favicon*/ $("link[rel='shortcut icon']").remove(); $("head").append("<link rel='shortcut icon' type='image/png' href='https://static.igem.org/mediawiki/2013/6/6d/Uppsalas-cow-con.png'>"); }); | ||
| + | </script> | ||
| + | |||
| + | <link href="https://2013.igem.org/Team:Uppsala/lightbox-css-code.css?action=raw&ctype=text/css" type="text/css" rel="stylesheet"> | ||
| + | |||
| + | <script src="https://2013.igem.org/Team:Uppsala/jquery-code.js?action=raw&ctype=text/javascript" type="text/javascript"></script> | ||
| + | |||
| + | <script src="https://2013.igem.org/Team:Uppsala/lightbox-code.js?action=raw&ctype=text/javascript" type="text/javascript"></script> | ||
</head> | </head> | ||
| Line 29: | Line 41: | ||
<div id="igem"><a href="https://2013.igem.org/Main_Page"><img class="igem" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_Uppsala_IGEM_log_blue.png"></a></div> | <div id="igem"><a href="https://2013.igem.org/Main_Page"><img class="igem" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_Uppsala_IGEM_log_blue.png"></a></div> | ||
| + | |||
| + | <img class="slogan" src="https://static.igem.org/mediawiki/2013/a/ad/Uppsala2013_Header.png"> | ||
</div> | </div> | ||
| Line 56: | Line 70: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/metabolic-engineering">Metabolic engineering</a> | <li><a href="https://2013.igem.org/Team:Uppsala/metabolic-engineering">Metabolic engineering</a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/p-coumaric-acid"> | + | <li><a href="https://2013.igem.org/Team:Uppsala/p-coumaric-acid">p-Coumaric acid</a></li> |
<li><a href="https://2013.igem.org/Team:Uppsala/resveratrol">Resveratrol</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/resveratrol">Resveratrol</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/lycopene">Lycopene</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/lycopene">Lycopene</a></li> | ||
| Line 77: | Line 91: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/modeling" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/6/63/Uppsala2013_Modeling.png"></a> | <li><a href="https://2013.igem.org/Team:Uppsala/modeling" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/6/63/Uppsala2013_Modeling.png"></a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/P-Coumaric-acid-pathway"> | + | <li><a href="https://2013.igem.org/Team:Uppsala/P-Coumaric-acid-pathway">Kinetic model</a></li> |
<li><a href="https://2013.igem.org/Team:Uppsala/modeling-tutorial">Modeling tutorial </a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/modeling-tutorial">Modeling tutorial </a></li> | ||
| + | |||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/toxicity-model">Toxicity model</a></li> | ||
</ul></li> | </ul></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/parts" id="list_type2"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/e/eb/Uppsala2013_parts.png"></a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/parts" id="list_type2"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/e/eb/Uppsala2013_parts.png"></a></li> | ||
| Line 87: | Line 103: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/carotenoid-group">Carotenoid group</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/carotenoid-group">Carotenoid group</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/chassi-group">Chassi group</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/chassi-group">Chassi group</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/advisors">Advisors</a></li> | ||
</ul></li> | </ul></li> | ||
| Line 96: | Line 113: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/public-opinion">Public opinion </a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/public-opinion">Public opinion </a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/Outreach">High school & media </a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/Outreach">High school & media </a></li> | ||
| - | + | <li><a href="https://2013.igem.org/Team:Uppsala/bioart">BioArt</a></li> | |
| + | <li><a href="https://2013.igem.org/Team:Uppsala/LactonutritiousWorld">A LactoWorld</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/killswitches">Killswitches</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/realization">Patent</a></li> | ||
</ul></li> | </ul></li> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a | + | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a></li> |
| - | + | ||
| - | + | ||
| - | + | ||
<li><a href="https://2013.igem.org/Team:Uppsala/notebook" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/3/36/Uppsala2013_Notebook.png"></a> | <li><a href="https://2013.igem.org/Team:Uppsala/notebook" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/3/36/Uppsala2013_Notebook.png"></a> | ||
<ul> | <ul> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/safety-form">Safety form</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/safety-form">Safety form</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/protocols">Protocols</a></li> | ||
</ul></li> | </ul></li> | ||
</ul> | </ul> | ||
| Line 113: | Line 131: | ||
<div id="main_content"> <!-- Put content here --> | <div id="main_content"> <!-- Put content here --> | ||
<h1 class="main-title">Miraculin</h1> | <h1 class="main-title">Miraculin</h1> | ||
| - | <h1>Sweet polypeptide goodness</h1> | + | <h1>Sweet polypeptide of goodness</h1> |
| - | Synsepalum Dulcificum is a plant found along the coast of West Africa. It grows red berries about the size of coffee beans that have a quite peculiar property: they make sour things taste sweet. This effect was first documented in the late 1700's, and has caused the berries to be referred to as "miracle berries". The "miraculous" property is actually caused by a glycoprotein dubbed miraculin, that binds to the taste receptors on the tongue and modifies their response. The exact mechanism of the interaction is not entirely understood, but it has been used as a natural sweetener in different circumstances throughout the centuries.<br><br> | + | Synsepalum Dulcificum is a plant found along the coast of West Africa. It grows red berries about the size of coffee beans that have a quite peculiar property: they make sour things taste sweet. This effect was first documented in the late 1700's, and has caused the berries to be referred to as "miracle berries". The "miraculous" property is actually caused by a glycoprotein dubbed miraculin, that binds to the taste receptors on the tongue and modifies their response. The exact mechanism of the interaction is not entirely understood, but it has been used as a natural sweetener in different circumstances throughout the centuries.<sup><a href="#ref_point">[1]</a><br><br> |
| - | <table class="miraculin_table"><tr><td><img class="miraculus_pic" src="https://static.igem.org/mediawiki/2013/1/11/Uppsala_miraculin_pic1.JPG"></td><td> | + | <table class="miraculin_table"><tr><td><a href="https://static.igem.org/mediawiki/2013/1/11/Uppsala_miraculin_pic1.JPG" data-lightbox="roadtrip"><img class="miraculus_pic" src="https://static.igem.org/mediawiki/2013/1/11/Uppsala_miraculin_pic1.JPG"></a></td><td> |
| - | <img class="miraculus_pic" src="https://static.igem.org/mediawiki/2013/9/95/Uppsala_miraculin_pic2.JPG"></td></tr></table> | + | <a href="https://static.igem.org/mediawiki/2013/9/95/Uppsala_miraculin_pic2.JPG" data-lightbox="roadtrip"><img class="miraculus_pic" src="https://static.igem.org/mediawiki/2013/9/95/Uppsala_miraculin_pic2.JPG"></a></td></tr></table> |
<i> </i> | <i> </i> | ||
The gene coding for the protein miraculin has never been used in iGEM before. Due to its unusual properties we decided to implement miraculin in our probiotic yoghurt where it would act as a form of sweetner <br><br> | The gene coding for the protein miraculin has never been used in iGEM before. Due to its unusual properties we decided to implement miraculin in our probiotic yoghurt where it would act as a form of sweetner <br><br> | ||
| - | < | + | <h1> Structure </h1> |
| + | <div> | ||
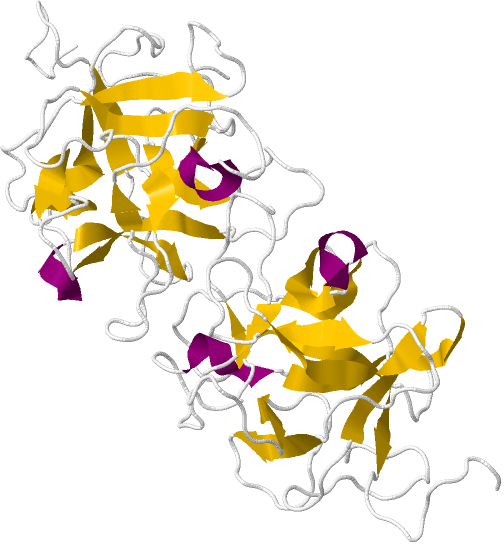
| + | <div> In its glycoprotein form, miraculin appears as a homotetramer, composed of two dimer subunits. The dimers, in turn, are composed of two monomers liked by disulfide bridge. The exact mechansm of interaction between miraculin and the taste-receptors is not completely explored, but it is known that miraculin changes it's shape at low pH-level, and that two histidine residues play an important role in binding to the receptor. This means that while miraculin can assocate to the receptors at normal circuimstances, it is not until the environment turns acidic that the conformation change that presumably causes the sweet sensation is initiated. When the pH returns to normal, so does the miraculin structure.<sup><a href="#ref_point">[2]</a> </div> | ||
| - | < | + | <div id="miraculin-box"> |
| - | < | + | <a href="https://static.igem.org/mediawiki/2013/6/62/Uppsala_miraculin_pic3.JPG" data-lightbox="roadtrip"><img class="miraculin-pic" src="https://static.igem.org/mediawiki/2013/6/62/Uppsala_miraculin_pic3.JPG"></a> |
| - | + | ||
| + | <div id="abcd"> | ||
| + | <i class="miraculin-text-left"><b>Figure 3.</b> the structure of the miraculin homodimer subunit. The 191 residues long polypeptide is glycosylated to a 13.9% carbohydrate content.</i> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div id="clear"></div> | ||
| + | |||
| + | </div> | ||
| + | <br><br><br> | ||
<h1>Methods</h1> | <h1>Methods</h1> | ||
We synthesized the miraculin gene along with a B0034 ribosome binding site for expression, the export tag USP45 for exportation of the complete glycopeptide out of the cell, and a 6xhis-tag for characterization purposes. Additionally, the gene was isolated with primers using biobrick overhangs for increased modularity in further use. We verify all our genetic constructs by sequencing<br><br> | We synthesized the miraculin gene along with a B0034 ribosome binding site for expression, the export tag USP45 for exportation of the complete glycopeptide out of the cell, and a 6xhis-tag for characterization purposes. Additionally, the gene was isolated with primers using biobrick overhangs for increased modularity in further use. We verify all our genetic constructs by sequencing<br><br> | ||
<h1>Results</h1> | <h1>Results</h1> | ||
| - | + | We were successful in both subcloning the synthetic construct to a desired plasmid and isolating the Miraculin gene by itself. The sequencencing was done at GATC biotch and Uppsala Genome center. We were not successful in either assembling or mutagenizing a promoter to the construct, and as such could not characterize it further. <br><br> | |
| - | We were successful in both subcloning the synthetic construct to a desired plasmid and isolating the | + | |
<h3>Biobricks</h3> | <h3>Biobricks</h3> | ||
<ul> | <ul> | ||
| Line 141: | Line 170: | ||
<li> <a href="http://parts.igem.org/Part:BBa_K1033121">BBa_K1033121</a> - Miraculin </li> | <li> <a href="http://parts.igem.org/Part:BBa_K1033121">BBa_K1033121</a> - Miraculin </li> | ||
</ul> | </ul> | ||
| - | + | <a name="ref_point"></a> | |
| + | <h1>References</h1> | ||
| + | <a>[1]</a> Slater, Joanna (2007-03-30). "To Make Lemons Into Lemonade, Try 'Miracle Fruit'" . Wall Street Journal. [2013-10-01].<br><br> | ||
| + | <a>[2]</a> Theerasilp S, Hitotsuya H, Nakajo S, Nakaya K, Nakamura Y, Kurihara Y (April 1989). "Complete amino acid sequence and structure characterization of the taste-modifying protein, miraculin". J. Biol. Chem. 264 (12): 6655–9. | ||
| + | < br><br> All pictures are taken from wikimedia commons | ||
</div> <!-- main_ontent ends --> | </div> <!-- main_ontent ends --> | ||
<div id="bottom-pic"> | <div id="bottom-pic"> | ||
| - | + | ||
</div> | </div> | ||
Latest revision as of 21:32, 28 October 2013
Miraculin
Sweet polypeptide of goodness
Synsepalum Dulcificum is a plant found along the coast of West Africa. It grows red berries about the size of coffee beans that have a quite peculiar property: they make sour things taste sweet. This effect was first documented in the late 1700's, and has caused the berries to be referred to as "miracle berries". The "miraculous" property is actually caused by a glycoprotein dubbed miraculin, that binds to the taste receptors on the tongue and modifies their response. The exact mechanism of the interaction is not entirely understood, but it has been used as a natural sweetener in different circumstances throughout the centuries.[1] |
 |
Structure
In its glycoprotein form, miraculin appears as a homotetramer, composed of two dimer subunits. The dimers, in turn, are composed of two monomers liked by disulfide bridge. The exact mechansm of interaction between miraculin and the taste-receptors is not completely explored, but it is known that miraculin changes it's shape at low pH-level, and that two histidine residues play an important role in binding to the receptor. This means that while miraculin can assocate to the receptors at normal circuimstances, it is not until the environment turns acidic that the conformation change that presumably causes the sweet sensation is initiated. When the pH returns to normal, so does the miraculin structure.[2]
Methods
We synthesized the miraculin gene along with a B0034 ribosome binding site for expression, the export tag USP45 for exportation of the complete glycopeptide out of the cell, and a 6xhis-tag for characterization purposes. Additionally, the gene was isolated with primers using biobrick overhangs for increased modularity in further use. We verify all our genetic constructs by sequencingResults
We were successful in both subcloning the synthetic construct to a desired plasmid and isolating the Miraculin gene by itself. The sequencencing was done at GATC biotch and Uppsala Genome center. We were not successful in either assembling or mutagenizing a promoter to the construct, and as such could not characterize it further.Biobricks
- BBa_K1033120 - B0034-ExpTag-Linker-Miraculin-Linker-6xHis
- BBa_K1033122 - B0034-Miraculin
- BBa_K1033121 - Miraculin
References
[1] Slater, Joanna (2007-03-30). "To Make Lemons Into Lemonade, Try 'Miracle Fruit'" . Wall Street Journal. [2013-10-01].[2] Theerasilp S, Hitotsuya H, Nakajo S, Nakaya K, Nakamura Y, Kurihara Y (April 1989). "Complete amino acid sequence and structure characterization of the taste-modifying protein, miraculin". J. Biol. Chem. 264 (12): 6655–9. < br>
All pictures are taken from wikimedia commons
 "
"