Team:Heidelberg/Tyrocidine week17 ms
From 2013.igem.org
| Line 1: | Line 1: | ||
| - | + | ||
== Final validation of BAP-I-cells transformed with pIK03 and pIK04 == | == Final validation of BAP-I-cells transformed with pIK03 and pIK04 == | ||
| Line 5: | Line 5: | ||
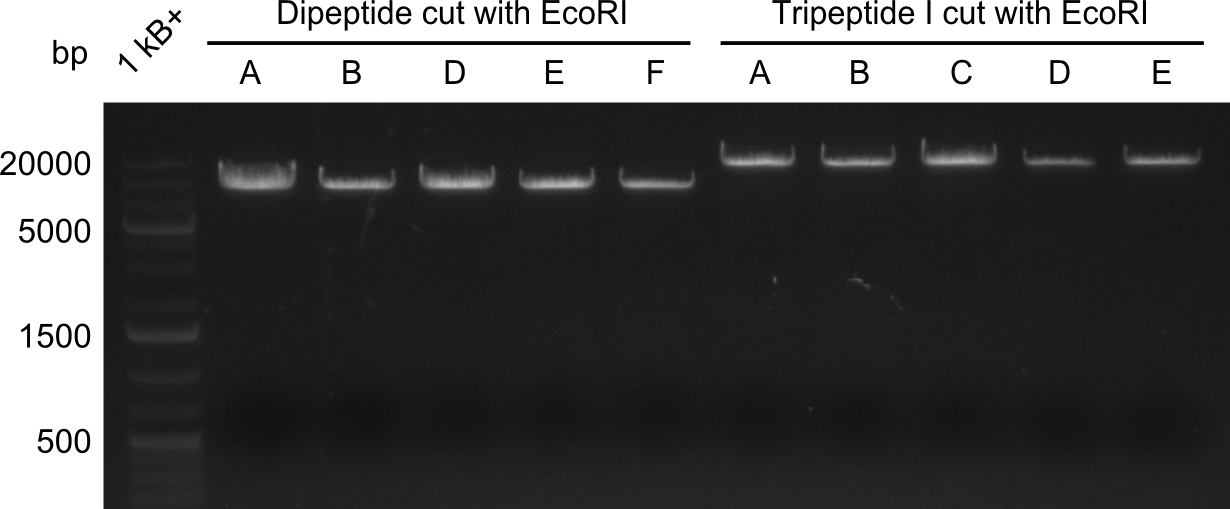
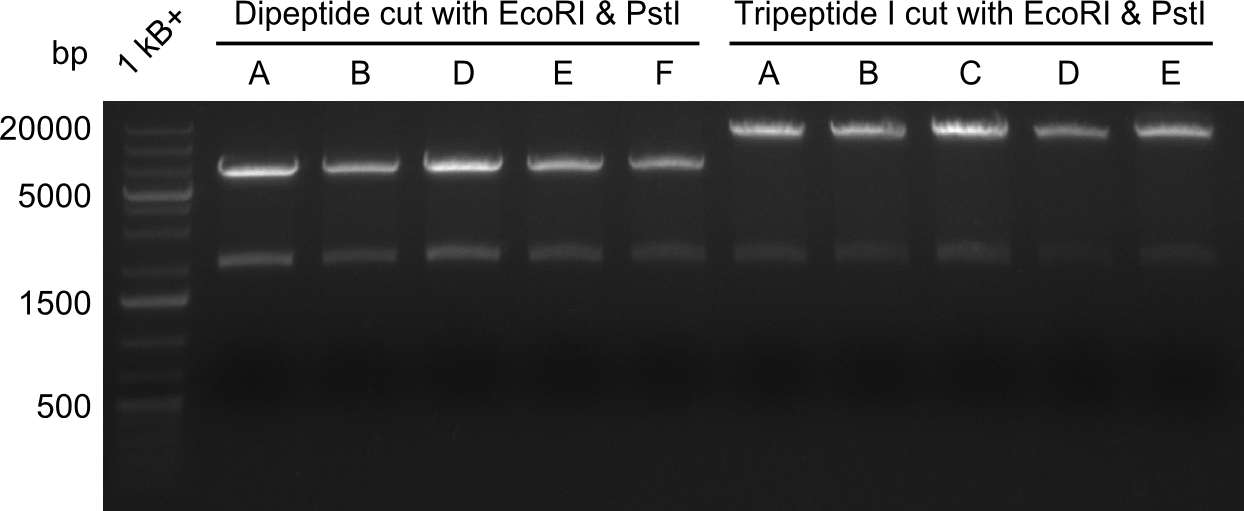
In order to select the samples that we will test on SDS-PAGE and in Mass-Spec, we decided to perform restriction digests and to send the positive samples to sequencing. Restriction digest was performed in a 20µl volume, using 2µl 10x CutSmart buffer, 1µl enzyme(s) and ~500ng DNA per reaction. In order to see both, the purity of the sample, as well as the correct sizes of insert and backbone, we digested every sample twice, once with EcoRI and PstI (in order to see, if the insert has the appropriate size) and once more with EcoRI alone (to see, if there is only one single band). At this stage, all plasmids were already transformed into our expression strain BAP-I. | In order to select the samples that we will test on SDS-PAGE and in Mass-Spec, we decided to perform restriction digests and to send the positive samples to sequencing. Restriction digest was performed in a 20µl volume, using 2µl 10x CutSmart buffer, 1µl enzyme(s) and ~500ng DNA per reaction. In order to see both, the purity of the sample, as well as the correct sizes of insert and backbone, we digested every sample twice, once with EcoRI and PstI (in order to see, if the insert has the appropriate size) and once more with EcoRI alone (to see, if there is only one single band). At this stage, all plasmids were already transformed into our expression strain BAP-I. | ||
| - | [[File: | + | [[File:Heidelberg_BAP_Di_Tri_Eco.png|300px]] |
| - | [[File: | + | [[File:Heidelberg_BAP_Di_Tri_EcoPst.png|300px]] |
| - | [[File: | + | [[File:Heidelberg_BAP_Di_EcoPst_Tri_EcoPst__DH10beta_Tet_Eco.png|300px]] |
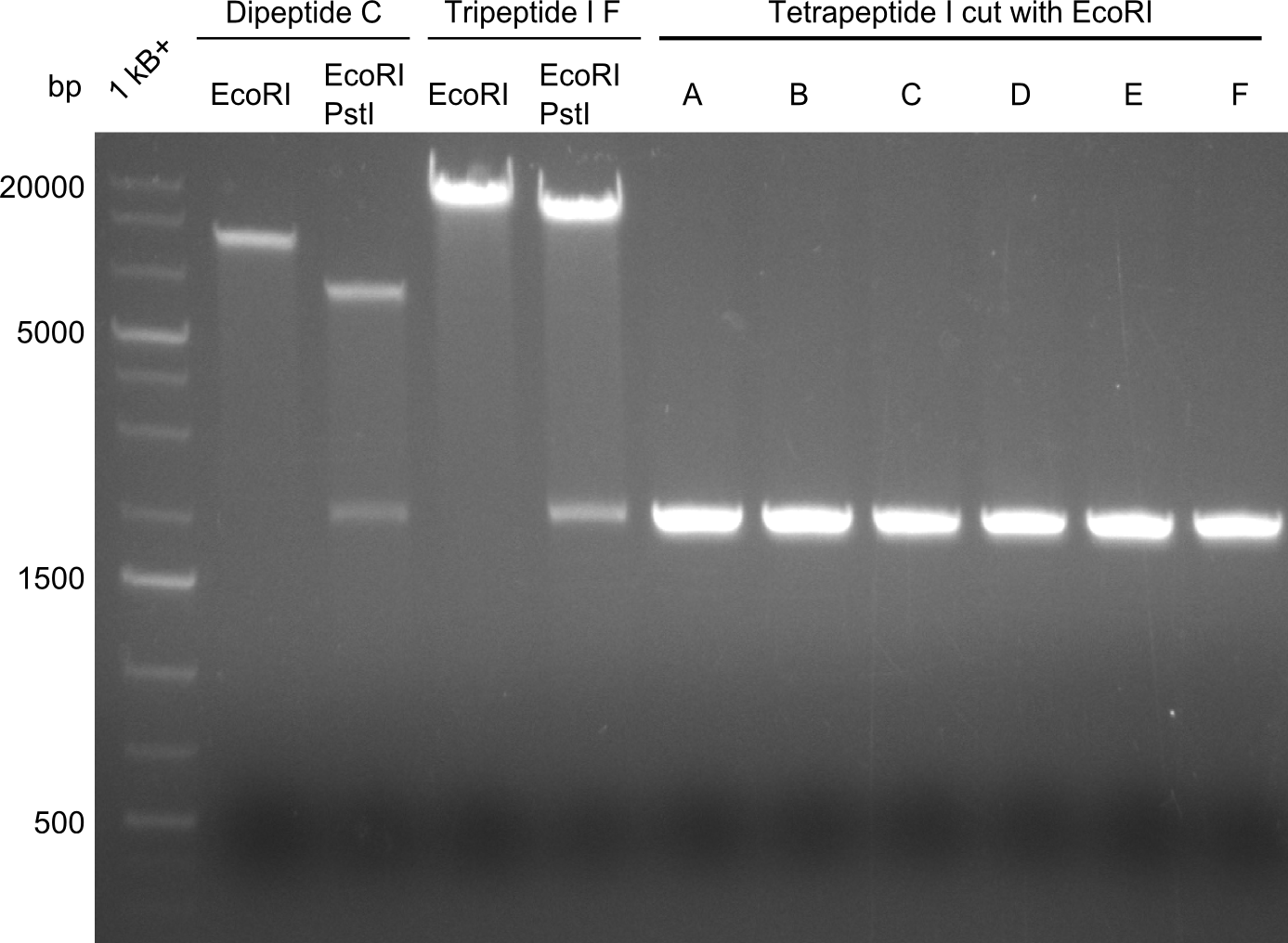
The restriction digests look very promising for the samples of the Dipeptide- and the Tripeptide-I-NRPS. We hence decided to use all of them on an SDS-PAGE. On the first two images, one can nicely see the expected cutting patterns. The third image was thought as comparison between the positive Di- and Tripeptide-NRPS samples and the Tetrapeptide-I-NRPS samples that we wanted to use. | The restriction digests look very promising for the samples of the Dipeptide- and the Tripeptide-I-NRPS. We hence decided to use all of them on an SDS-PAGE. On the first two images, one can nicely see the expected cutting patterns. The third image was thought as comparison between the positive Di- and Tripeptide-NRPS samples and the Tetrapeptide-I-NRPS samples that we wanted to use. | ||
| Line 26: | Line 26: | ||
Unfortunately, the restriction digest described above did not reveal the expected and desired fragment sizes: | Unfortunately, the restriction digest described above did not reveal the expected and desired fragment sizes: | ||
| - | [[File: | + | [[File:Heidelberg_BAP_Di_EcoPst_Tri_EcoPst__DH10beta_Tet_Eco.png|300px]] |
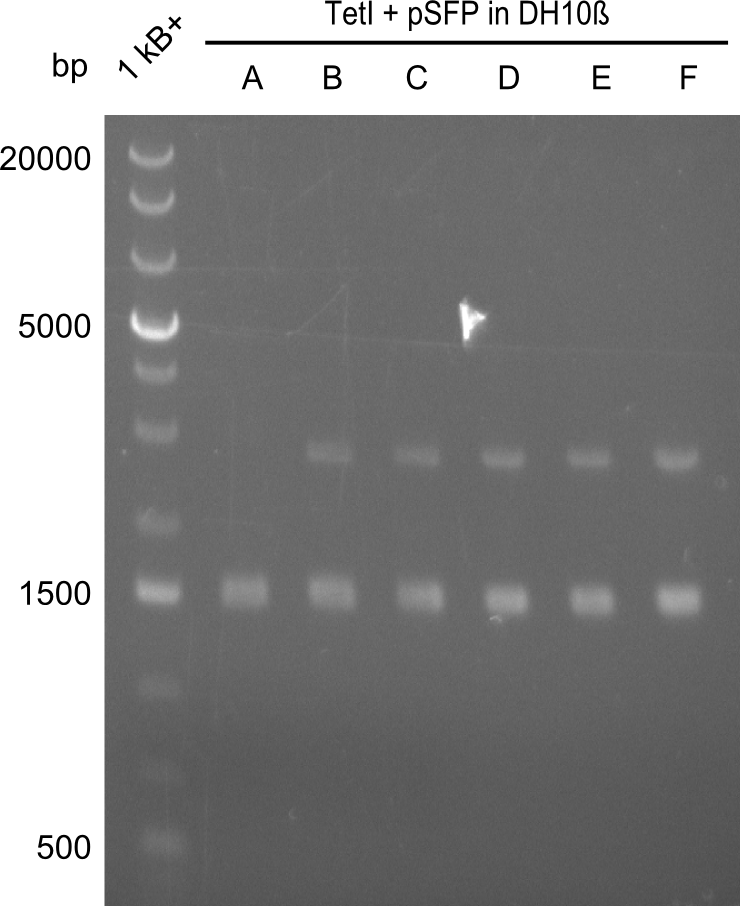
At first, we believed that the chemically competent cells could not incorporate the 16kb plasmid in a heat-shock transformation, therefore we started a Co-transformation of pPW01 (Tetrapeptide I) and the PPTase sfp so that we could "stay" in DH10β-cells where we had electrocompetent cells available. | At first, we believed that the chemically competent cells could not incorporate the 16kb plasmid in a heat-shock transformation, therefore we started a Co-transformation of pPW01 (Tetrapeptide I) and the PPTase sfp so that we could "stay" in DH10β-cells where we had electrocompetent cells available. | ||
| - | [[File: | + | [[File:Heidelberg_DH10beta_TetI_pSFP_Eco.png|300px]] |
Even though screening by restriction digest is not that easy when two plasmids are present, it is clear that the approach did not work and that there has to be another problem. | Even though screening by restriction digest is not that easy when two plasmids are present, it is clear that the approach did not work and that there has to be another problem. | ||
Latest revision as of 16:43, 4 October 2013
Contents |
Final validation of BAP-I-cells transformed with pIK03 and pIK04
Restriction Digests
In order to select the samples that we will test on SDS-PAGE and in Mass-Spec, we decided to perform restriction digests and to send the positive samples to sequencing. Restriction digest was performed in a 20µl volume, using 2µl 10x CutSmart buffer, 1µl enzyme(s) and ~500ng DNA per reaction. In order to see both, the purity of the sample, as well as the correct sizes of insert and backbone, we digested every sample twice, once with EcoRI and PstI (in order to see, if the insert has the appropriate size) and once more with EcoRI alone (to see, if there is only one single band). At this stage, all plasmids were already transformed into our expression strain BAP-I.
The restriction digests look very promising for the samples of the Dipeptide- and the Tripeptide-I-NRPS. We hence decided to use all of them on an SDS-PAGE. On the first two images, one can nicely see the expected cutting patterns. The third image was thought as comparison between the positive Di- and Tripeptide-NRPS samples and the Tetrapeptide-I-NRPS samples that we wanted to use.
SDS-PAGE
Recipe for Running Buffer:
- 100 mM Tris-HCl
- 4% SDS
- 0.2% bromophenol-blue
- 20% glycerol
- 200 mM DTT
Unfortunately there were no bands showing up on the gel, which is most probably due to a wrong pH of the loading buffer. Hence this was changed for further optimization.
Tetrapeptide I
Unfortunately, the restriction digest described above did not reveal the expected and desired fragment sizes:
At first, we believed that the chemically competent cells could not incorporate the 16kb plasmid in a heat-shock transformation, therefore we started a Co-transformation of pPW01 (Tetrapeptide I) and the PPTase sfp so that we could "stay" in DH10β-cells where we had electrocompetent cells available.
Even though screening by restriction digest is not that easy when two plasmids are present, it is clear that the approach did not work and that there has to be another problem.
 "
"