Team:Uppsala/reporter-genes
From 2013.igem.org
Sabrijamal (Talk | contribs) |
|||
| (32 intermediate revisions not shown) | |||
| Line 13: | Line 13: | ||
</style> | </style> | ||
| + | |||
| + | <script type="text/javascript"> | ||
| + | /*Acts once the document is loaded*/ $(document).ready(function() { /*Move the tn-main-wrap as child of <body>*/ $("#tn-main-wrap-wrap").prependTo($("body")); /*Move the menubars in the tn-main-wrap*/ $("#menubar.left-menu").appendTo($("#tn-main-wrap")); $("#menubar.right-menu").appendTo($("#tn-main-wrap")); | ||
| + | /*Spoiler JS*/ $(".tn-spoiler div").slideUp(); $(".tn-spoiler a").click(function(e) { e.preventDefault(); $(".tn-spoiler-active").removeClass("tn-spoiler-active"); $(this).parent().addClass("tn-spoiler-active"); $(".tn-spoiler").not(".tn-spoiler-active").children("div").slideUp(); $(".tn-spoiler-active").children("div").slideToggle(); }); | ||
| + | /*Add favicon*/ $("link[rel='shortcut icon']").remove(); $("head").append("<link rel='shortcut icon' type='image/png' href='https://static.igem.org/mediawiki/2013/6/6d/Uppsalas-cow-con.png'>"); }); | ||
| + | </script> | ||
| + | |||
| + | <link href="https://2013.igem.org/Team:Uppsala/lightbox-css-code.css?action=raw&ctype=text/css" type="text/css" rel="stylesheet"> | ||
| + | |||
| + | <script src="https://2013.igem.org/Team:Uppsala/jquery-code.js?action=raw&ctype=text/javascript" type="text/javascript"></script> | ||
| + | |||
| + | <script src="https://2013.igem.org/Team:Uppsala/lightbox-code.js?action=raw&ctype=text/javascript" type="text/javascript"></script> | ||
</head> | </head> | ||
| Line 59: | Line 71: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/metabolic-engineering">Metabolic engineering</a> | <li><a href="https://2013.igem.org/Team:Uppsala/metabolic-engineering">Metabolic engineering</a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/p-coumaric-acid"> | + | <li><a href="https://2013.igem.org/Team:Uppsala/p-coumaric-acid">p-Coumaric acid</a></li> |
<li><a href="https://2013.igem.org/Team:Uppsala/resveratrol">Resveratrol</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/resveratrol">Resveratrol</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/lycopene">Lycopene</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/lycopene">Lycopene</a></li> | ||
| Line 80: | Line 92: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/modeling" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/6/63/Uppsala2013_Modeling.png"></a> | <li><a href="https://2013.igem.org/Team:Uppsala/modeling" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/6/63/Uppsala2013_Modeling.png"></a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/P-Coumaric-acid-pathway"> | + | <li><a href="https://2013.igem.org/Team:Uppsala/P-Coumaric-acid-pathway">Kinetic model</a></li> |
<li><a href="https://2013.igem.org/Team:Uppsala/modeling-tutorial">Modeling tutorial </a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/modeling-tutorial">Modeling tutorial </a></li> | ||
| + | |||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/toxicity-model">Toxicity model</a></li> | ||
</ul></li> | </ul></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/parts" id="list_type2"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/e/eb/Uppsala2013_parts.png"></a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/parts" id="list_type2"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/e/eb/Uppsala2013_parts.png"></a></li> | ||
| Line 90: | Line 104: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/carotenoid-group">Carotenoid group</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/carotenoid-group">Carotenoid group</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/chassi-group">Chassi group</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/chassi-group">Chassi group</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/advisors">Advisors</a></li> | ||
</ul></li> | </ul></li> | ||
| Line 99: | Line 114: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/public-opinion">Public opinion </a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/public-opinion">Public opinion </a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/Outreach">High school & media </a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/Outreach">High school & media </a></li> | ||
| - | + | <li><a href="https://2013.igem.org/Team:Uppsala/bioart">BioArt</a></li> | |
| + | <li><a href="https://2013.igem.org/Team:Uppsala/LactonutritiousWorld">A LactoWorld</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/killswitches">Killswitches</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/realization">Patent</a></li> | ||
</ul></li> | </ul></li> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a | + | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a></li> |
| - | + | ||
| - | + | ||
| - | + | ||
<li><a href="https://2013.igem.org/Team:Uppsala/notebook" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/3/36/Uppsala2013_Notebook.png"></a> | <li><a href="https://2013.igem.org/Team:Uppsala/notebook" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/3/36/Uppsala2013_Notebook.png"></a> | ||
<ul> | <ul> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/safety-form">Safety form</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/safety-form">Safety form</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/protocols">Protocols</a></li> | ||
</ul></li> | </ul></li> | ||
</ul> | </ul> | ||
| Line 124: | Line 140: | ||
<p>One of the most fundamental parts required in a new chassi is a reporter gene. Unfortunately, some fluorescent proteins do not work well in Lactobacillus, possibly due to lower levels of internal pH in the cytosol. | <p>One of the most fundamental parts required in a new chassi is a reporter gene. Unfortunately, some fluorescent proteins do not work well in Lactobacillus, possibly due to lower levels of internal pH in the cytosol. | ||
<br><br> | <br><br> | ||
| - | <p>One popular fluorescent protein that does work is mCherry. We have created a biobricked version of mCherry, <a href="http://parts.igem.org/Part:BBa_K1033250"> BBa_K1033250 </a> codon optimized for Lactobacillus reuteri, however it should work well in most species in the Lactobacillus genus. The gene has been modified in accordance with the Freiburgh fusion standard 25. Because of time constraints we have not yet had time to characterise it but it has been used in non-biobrick format by other researchers with positive results</p> | + | <p>One popular fluorescent protein that does work is <a href="http://parts.igem.org/Part:BBa_K1033250">mCherry.</a> We have created a biobricked version of mCherry, <a href="http://parts.igem.org/Part:BBa_K1033250"> BBa_K1033250 </a> codon optimized for Lactobacillus reuteri, however it should work well in most species in the Lactobacillus genus. The gene has been modified in accordance with the Freiburgh fusion standard 25. Because of time constraints we have not yet had time to characterise it but it has been used in non-biobrick format by other researchers with positive results</p> |
<h3>Prefix</h3> | <h3>Prefix</h3> | ||
| Line 136: | Line 152: | ||
<p>There are many things we take for granted when working with bacteria such as E. coli. An initial problem when learning to work with Lactobacillus is to simply be able to do successful transformations of plasmids, something that is more difficult and cumbersome to do compared to E. coli. To see if you have been able to master a transformation with Lactobacillus some basic reporter genes that reflects visible light makes the validation process a lot easier.</p> | <p>There are many things we take for granted when working with bacteria such as E. coli. An initial problem when learning to work with Lactobacillus is to simply be able to do successful transformations of plasmids, something that is more difficult and cumbersome to do compared to E. coli. To see if you have been able to master a transformation with Lactobacillus some basic reporter genes that reflects visible light makes the validation process a lot easier.</p> | ||
| - | <img class="method-plasmid1" src="https://static.igem.org/mediawiki/2013/9/9b/Uppsala2013_reporter-plasmid.jpg"> | + | <a href="https://static.igem.org/mediawiki/2013/9/9b/Uppsala2013_reporter-plasmid.jpg" data-lightbox="roadtrip"><img class="method-plasmid1" src="https://static.igem.org/mediawiki/2013/9/9b/Uppsala2013_reporter-plasmid.jpg"></a> |
| - | <p>Above is a picture of one of our chromo proteins and RBS fused together with two of our synthetic promoters, <a href="http://parts.igem.org/Part:BBa_K1033221"> CP11 </a> and <a href="http://parts.igem.org/Part:BBa_K1033221"> CP29 </a>. These promoters have been characterised and has been shown to work both in E. coli and Lactobacillus. We wanted to see if our chromoproteins could work as visual reporters. The construct has been shown to work fine with our shuttle vector in E. coli as can clearly be seen in the picture below with blue bacterial colonies. We have however not yet been able to transform it into Lactobacillus.</p> | + | <p>Above is a picture of one of our chromo proteins and RBS fused together with two of our synthetic promoters, <a href="http://parts.igem.org/Part:BBa_K1033221"> CP11 </a> and <a href="http://parts.igem.org/Part:BBa_K1033221"> CP29 </a>. These promoters have been characterised and has been shown to work both in E. coli and Lactobacillus. We wanted to see if our chromoproteins could work as visual reporters. The construct has been shown to work fine with our shuttle vector in E. coli as can clearly be seen in the picture below with blue bacterial colonies. We have however not yet been able to transform it into Lactobacillus. Below in the picture is a plate with our shuttle vector <a href="http://parts.igem.org/Part:BBa_K1033206">BBa_K1033206</a> containing the chromoprotein amilCP fused with the synthetic promoter CP29 from our promoter collection. The colonies are clearly blue efter 24 hours in E. coli.</p> |
| + | <br> | ||
| - | <img class="shuttle_vec" src=" | + | <a href="https://static.igem.org/mediawiki/2013/b/ba/Uppsala2013_Shuttle_Vector_pSBLBC_cp29_amilCP.JPG" data-lightbox="roadtrip" title="Above in the picture is a plate with our shuttle vector BBa_K1033206 containing the chromoprotein amilCP fused with the synthetic promoter CP29 from our promoter collection. The colonies are clearly blue efter 24 hours in E. coli."><img class="shuttle_vec" src="https://static.igem.org/mediawiki/2013/1/18/Uppsala2013_Shuttle_Vector_pSBLBC_cp29_amilCP1.png"></a> |
| - | "> </ | + | |
| + | |||
| + | <!-- | ||
| + | <div id="shuttle_vec_cont"> | ||
| + | |||
| + | </div> | ||
| + | --> | ||
<h1> Biobricks </h1> | <h1> Biobricks </h1> | ||
| Line 153: | Line 176: | ||
<div id="bottom-pic"> | <div id="bottom-pic"> | ||
| - | + | ||
</div> | </div> | ||
Latest revision as of 23:22, 28 October 2013
Reporter genes

One of the most fundamental parts required in a new chassi is a reporter gene. Unfortunately, some fluorescent proteins do not work well in Lactobacillus, possibly due to lower levels of internal pH in the cytosol.
One popular fluorescent protein that does work is mCherry. We have created a biobricked version of mCherry, BBa_K1033250 codon optimized for Lactobacillus reuteri, however it should work well in most species in the Lactobacillus genus. The gene has been modified in accordance with the Freiburgh fusion standard 25. Because of time constraints we have not yet had time to characterise it but it has been used in non-biobrick format by other researchers with positive results
Prefix
gaattccgcggccgcttctagatggccggc...
Suffix
... accggttaatactagtagcggccgctgcag
Chromoproteins in Lactobacillus
There are many things we take for granted when working with bacteria such as E. coli. An initial problem when learning to work with Lactobacillus is to simply be able to do successful transformations of plasmids, something that is more difficult and cumbersome to do compared to E. coli. To see if you have been able to master a transformation with Lactobacillus some basic reporter genes that reflects visible light makes the validation process a lot easier.

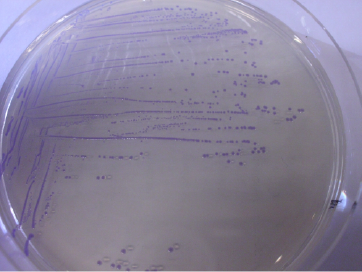
Above is a picture of one of our chromo proteins and RBS fused together with two of our synthetic promoters, CP11 and CP29 . These promoters have been characterised and has been shown to work both in E. coli and Lactobacillus. We wanted to see if our chromoproteins could work as visual reporters. The construct has been shown to work fine with our shuttle vector in E. coli as can clearly be seen in the picture below with blue bacterial colonies. We have however not yet been able to transform it into Lactobacillus. Below in the picture is a plate with our shuttle vector BBa_K1033206 containing the chromoprotein amilCP fused with the synthetic promoter CP29 from our promoter collection. The colonies are clearly blue efter 24 hours in E. coli.
Biobricks
BBa_K1033250 - mCherry fusion protein codon optimised for Lactobacillus reuteriBBa_K1033280 - aeBlue blue chromoprotein with strong promoter
BBa_K1033281 - amilCP blue chromoprotein with medium strength promoter
BBa_K1033282 - amilCP blue chromoprotein with strong promoter
 "
"










