Team:TU-Munich/Modeling/Kill Switch
From 2013.igem.org
ChristopherW (Talk | contribs) (→Stochastic Model) |
|||
| (16 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
==Kill Switch Modeling== | ==Kill Switch Modeling== | ||
| - | + | <div> | |
| - | ===Purpose=== | + | ====Purpose==== |
The idea of our [https://2013.igem.org/Team:TU-Munich/Project/Killswitch kill switch] is to kill off our moss, as soon as it leaves the filter system. For this purpose two methods were proposed: | The idea of our [https://2013.igem.org/Team:TU-Munich/Project/Killswitch kill switch] is to kill off our moss, as soon as it leaves the filter system. For this purpose two methods were proposed: | ||
| Line 15: | Line 15: | ||
To decide between these two methods we modelled the vitality V of the cell (a number between 0 and 1, so a perfectly functional cell has V=1, a dead cell V=0) and depending on the tested method the concentration of siRNA R and nuclease N as appropriate. Both concentrations are normalized to the unit interval [0,1]. | To decide between these two methods we modelled the vitality V of the cell (a number between 0 and 1, so a perfectly functional cell has V=1, a dead cell V=0) and depending on the tested method the concentration of siRNA R and nuclease N as appropriate. Both concentrations are normalized to the unit interval [0,1]. | ||
| - | |||
==siRNA Model== | ==siRNA Model== | ||
| Line 83: | Line 82: | ||
This result makes intuitive sense, because as the function of the cell is repressed the cell produces less of the inhibiting siRNA, which leads to a regeneration of the cell. Eventually a steady state at a lower vitality is reached, where the vitality stays constant. | This result makes intuitive sense, because as the function of the cell is repressed the cell produces less of the inhibiting siRNA, which leads to a regeneration of the cell. Eventually a steady state at a lower vitality is reached, where the vitality stays constant. | ||
| - | === Stochastic Model === | + | ==== Stochastic Model ==== |
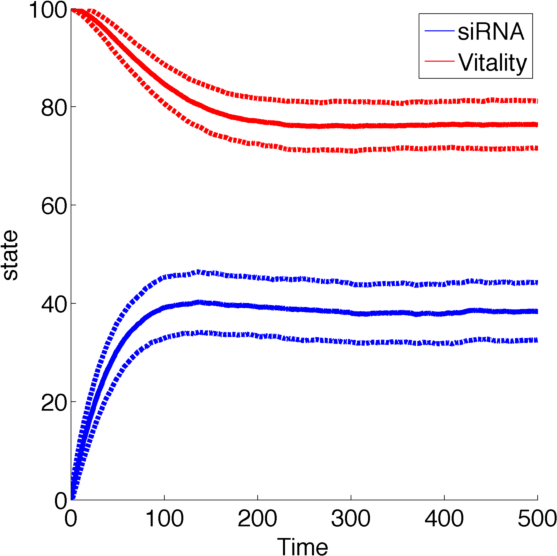
| - | [[File:TUM13_SiRNA_stoch.png|thumb|right|400px|Averaging over 1000 realisations of the Gillespie algorithm. Solid line indicates the mean, dashed lines indicate the 1-σ interval.]] | + | [[File:TUM13_SiRNA_stoch.png|thumb|right|400px| '''Figure 2''': Averaging over 1000 realisations of the Gillespie algorithm. Solid line indicates the mean, dashed lines indicate the 1-σ interval.]] |
| - | As number of siRNA in a cell can be very small, we cannot always assume that there is an sufficient amount of RNA such we can rule out stochastic effect. To deal with this problem we set up an analogous stochastic model constisting of | + | As number of siRNA in a cell can be very small, we cannot always assume that there is an sufficient amount of RNA such we can rule out stochastic effect. To deal with this problem we set up an analogous stochastic model constisting of a Markov Jump Process of the two species with a total of 4 reactions. |
| + | |||
| + | [[File:TUM13_SiRNA_stoch_system.png|center|400px]] | ||
| - | |||
The state space [0,1] was discretised to [1,2,3,...,100] for both species R and N while the reaction rates were kept the same. | The state space [0,1] was discretised to [1,2,3,...,100] for both species R and N while the reaction rates were kept the same. | ||
| Line 95: | Line 95: | ||
To simulate the model we used and implementation of the Gillespie Algorithm with 1000 realisations and then averaged over all realisations. | To simulate the model we used and implementation of the Gillespie Algorithm with 1000 realisations and then averaged over all realisations. | ||
| - | With this implementation the qualitative behaviour of the system remains unchanged, while the quantitative | + | With this implementation the qualitative behaviour of the system remains unchanged, while the quantitative changes which is due to the discretization. |
==Nuclease Model== | ==Nuclease Model== | ||
====Governing equations==== | ====Governing equations==== | ||
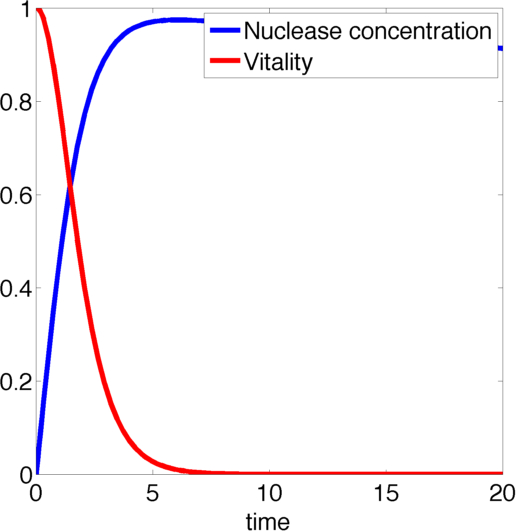
| - | [[File:TUM13_nuc_graph.png|thumb|right|400px| '''Figure | + | [[File:TUM13_nuc_graph.png|thumb|right|400px| '''Figure 3:''' Example solution to our nuclease model with parameters p<sub>1</sub>=1, p<sub>3</sub>=0.5, p<sub>4</sub>=0.005.]] |
We determined the governing equations of this model to be the following:[[File:TUM13_nuc_formula.png|center]] | We determined the governing equations of this model to be the following:[[File:TUM13_nuc_formula.png|center]] | ||
| Line 287: | Line 287: | ||
</nowiki> | </nowiki> | ||
| - | ==== | + | ====Nuclease model script==== |
<nowiki>% f = @(N,V,k) [p(3)* V - p(4) * N , -p(1) * N * V ]; | <nowiki>% f = @(N,V,k) [p(3)* V - p(4) * N , -p(1) * N * V ]; | ||
| Line 309: | Line 309: | ||
export_fig nuc_pic.png</nowiki> | export_fig nuc_pic.png</nowiki> | ||
| - | + | </div> | |
<!-- Ende des Inhalts --> | <!-- Ende des Inhalts --> | ||
</div> | </div> | ||
Latest revision as of 02:03, 29 October 2013
Kill Switch Modeling
Purpose
The idea of our kill switch is to kill off our moss, as soon as it leaves the filter system. For this purpose two methods were proposed:
- siRNA method: When some trigger is activated, siRNA is expressed inhibiting the expression of a vital gene
- nuclease method: When some trigger is activated, a nuclease is released destroying the DNA of the cell
To decide between these two methods we modelled the vitality V of the cell (a number between 0 and 1, so a perfectly functional cell has V=1, a dead cell V=0) and depending on the tested method the concentration of siRNA R and nuclease N as appropriate. Both concentrations are normalized to the unit interval [0,1].
siRNA Model
Governing equations
We determined the governing equations of this model to be the following:with initial conditions V(0) = 1 and R(0) = 0, where at the time t=0 the trigger is activated.
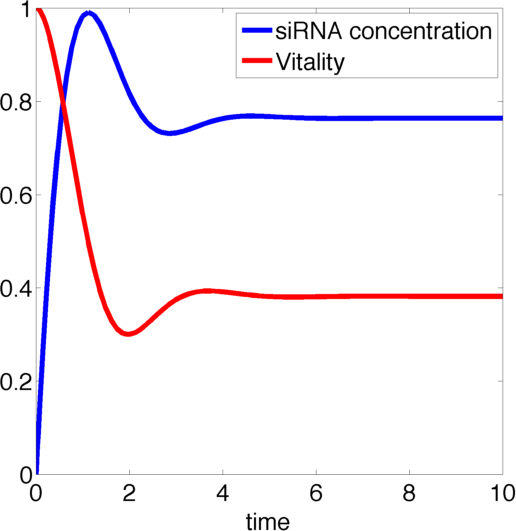
Figure 1, created by our MatLab script siRNA model shown below, on the right shows a solution to this system for some example parameters. In this case the vitality of the cell decreases to somewhere around 0.3 while at the same time after an initial peak the siRNA concentration also drops upon which the vitality settles in at about 0.4.
So it appears, that the siRNA approach does not achieve the required death of the cells. In the following it will be shown, that this is the case for all parameter values.
Calculation of stable points and analysis
The steady points V* and R* of this system have to satisfy Defining
If α = 1, the unique steady point is ![]() .
.
These are both negative, so this is a stable point, i.e. an attractor.
Now only one of these is in the sensible range, because
So there is only one steady point in this range, namely:
By expanding the fraction by ![]() we can rewrite V* as
we can rewrite V* as
So using the results from above, we get: ![]()
Defining ![]() , the eigenvalues are given by
, the eigenvalues are given by
So ![]() , which means that this is a stable attractor.
, which means that this is a stable attractor.
Result of our model
So for the siRNA there always is a stable point for the vitality between (but excluding) 0 and 1, so the moss is not killed-off completely, just impeded in its growth.
Interpretation
This result makes intuitive sense, because as the function of the cell is repressed the cell produces less of the inhibiting siRNA, which leads to a regeneration of the cell. Eventually a steady state at a lower vitality is reached, where the vitality stays constant.
Stochastic Model
As number of siRNA in a cell can be very small, we cannot always assume that there is an sufficient amount of RNA such we can rule out stochastic effect. To deal with this problem we set up an analogous stochastic model constisting of a Markov Jump Process of the two species with a total of 4 reactions.
The state space [0,1] was discretised to [1,2,3,...,100] for both species R and N while the reaction rates were kept the same.
To simulate the model we used and implementation of the Gillespie Algorithm with 1000 realisations and then averaged over all realisations.
With this implementation the qualitative behaviour of the system remains unchanged, while the quantitative changes which is due to the discretization.
Nuclease Model
Governing equations
We determined the governing equations of this model to be the following:with initial conditions V(0) = 1 and N(0) = 0, where at the time t=0 the trigger is activated.
Figure 2, created by our MatLab script nuclease model shown below, on the left shows a solution to this system for some example parameters. It clearly shows that in this case the vitality of the cell decreases to 0 and remains there, i.e. that the cell has died.
In the following we will verify, that this is the case for all parameter values.
Calculation of stable points and analysis
Any steady state given by V* and N* of this system must to satisfy
The eigenvector corresponding to the zero eigenvalue is ![]() . It is obvious from the equations that any disturbance away from the steady state along this vector will decay back to the steady state, so this is a stable attractor.
. It is obvious from the equations that any disturbance away from the steady state along this vector will decay back to the steady state, so this is a stable attractor.
Results of our model
The nuclease always reduces the vitality of the moss to 0, i.e. kills it off completely.
Interpretation
Again this result is very intuitive, since the moss cannot regenerate from the destruction of its genome.
Conclusions
For a functional kill-switch it is necessary, that the cells are actually killed and not just live on with reduced vitality. So based on our modeling results the siRNA approach is not satisfactory, while the nuclease satisfies the requirement. As a result the team pursued the nuclease approach leading to our final kill-switch.
MatLab Scripts
siRNA model script
% f = @(R,V,k) [k(3)* V - k(4) * R , -k(1) * R * V + k(2) *(V -1)* (R -1)];
g = @(k,y) [k(3)* y(2)- k(4)*y(1); -k(1)*y(1)*y(2)+k(2)*(y(2)-1)*(y(1)-1)];
%k= [ k1, k2, k3, k4 ]; %insert the appropriate reaction rate constant
k = [ 1, 2, 2, 1];
[TOUT, YOUT] = ode45(@(t,y) g(k,y) ,[0,10],[0;1]);
plot(TOUT, YOUT(:,1), 'markersize', 15, 'linewidth', 5)
hold on; plot(TOUT, YOUT(:,2), 'r', 'markersize', 15, 'linewidth', 5); hold off
legend('siRNA concentration','Vitality');
xlabel('time');
set(gca,'FontSize',24);
set(gcf,'position', [100 100 600 600]);
axis square
set(gcf,'Color','w');
export_fig siRNA_pic.png
Stochastic siRNA script
%% 1 States
% [R,V]
x0 = [0,100];
%% 2 Reactions
% stoichiometric matrix
S = zeros(4,2);
%% 2.1 increase siRNA
% R -> R + 1
% rate k3 * V
S(1,:) = [1,0];
%% 2.2 degrade siRNA
% R -> R - 1
% rate k4 * R
S(2,:) = [-1,0];
%% 2.3 increase vitality
% V -> V + 1
% rate k2 * ( V - 1 ) * ( R - 1 )
S(3,:) = [0,1];
%% 2.4 decrease vitality
% V -> V - 1
% rate k1 * R * V
S(4,:) = [0,-1];
param = [ 1, 2, 1, 2];
% rates
acell = { @(t,x,k) k(3) * x(2)/100,
@(t,x,k) k(4) * x(1)/100,
@(t,x,k) k(2) * ( x(2) - 100 )/100 * ( x(1) - 100 )/100,
@(t,x,k) k(1) * x(1) * x(2)/100};
a = @(t,x,k) [feval(acell{1},t,x,k),feval(acell{2},t,x,k),feval(acell{3},t,x,k),feval(acell{4},t,x,k)];
% time vector
N_time = 100;
tt = linspace(0,100,N_time);
% number of runs
N_repeat = 1000;
% output
X_runs = zeros(N_repeat,N_time,length(x0));
%plotting
figure(1)
clf
hold on
for j = 1:N_repeat
[X_runs(j,:,:)]=SSA(x0,S,a,tt,param);
end
figure
hold on
plot(tt,mean(X_runs(:,:,1)),'b')
plot(tt,mean(X_runs(:,:,2)),'r')
legend('siRNA','Vitality')
for k = 1:length(x0)
switch k
case 1
plot(tt,mean(X_runs(:,:,k)),'b')
plot(tt,mean(X_runs(:,:,k))+sqrt(var(X_runs(:,:,k))),'--b')
plot(tt,mean(X_runs(:,:,k))-sqrt(var(X_runs(:,:,k))),'--b')
case 2
plot(tt,mean(X_runs(:,:,k)),'r')
plot(tt,mean(X_runs(:,:,k))+sqrt(var(X_runs(:,:,k))),'--r')
plot(tt,mean(X_runs(:,:,k))-sqrt(var(X_runs(:,:,k))),'--r')
end
end
find(X_runs(:,:,2)==0,1,'first')
set(gcf,'Color','w')
axis square
export_fig siRNA_stochastic.png
Gillespie
function [ xx, XX, TT ] = SSA( x0, S, a, tt , param)
%SSA Summary of this function goes here
% Detailed explanation goes here
% initialise
TT(1) = 0;
XX(1,:) = x0;
% step
i = 1;
while (TT(end)<tt(end));
% increment step
i = i + 1;
%compute rates
at = a(TT(i-1),XX(i-1,:),param);
a0 = sum(at);
for k = 1 : length(at)
aj(k) = sum(at(1:k)/a0);
end
%sample time
TT(i) = TT(i-1) + expinv(rand,1/a0);
% select reaction that happens
j = find(aj>rand,1,'first');
% add reaction
XX(i,:) = XX(i-1,:) + S(j,:);
end
xx = zeros(length(tt),length(x0));
for j = 1:length(tt)
xx(j,:) = XX(find(TT<=tt(j),1,'last'),:);
end
end
Nuclease model script
% f = @(N,V,k) [p(3)* V - p(4) * N , -p(1) * N * V ];
g = @(p,y) [p(3)* y(2)- p(4)*y(1); -p(1)*y(1)*y(2)];
%p= [ p1, p2, p3, p4 ]; %insert the appropriate reaction rate constant
p = [ 1, NaN, 1, 0.5 ];
[TOUT, YOUT] = ode45(@(t,y) g(p,y) ,[0,20],[0;1]);
plot(TOUT, YOUT(:,1), 'markersize', 15, 'linewidth', 5)
hold on; plot(TOUT, YOUT(:,2), 'r', 'markersize', 15, 'linewidth', 5); hold off
legend('Nuclease concentration','Vitality')
xlabel('time');
set(gca,'FontSize',24);
set(gcf,'position', [100 100 600 600]);
axis square
set(gcf,'Color','w')
export_fig nuc_pic.png
 "
"



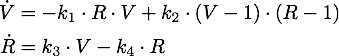
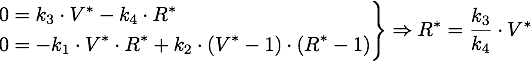
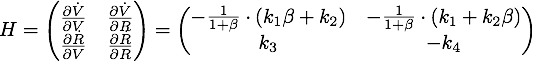
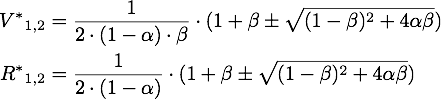
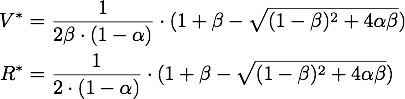
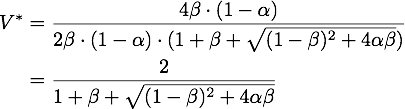
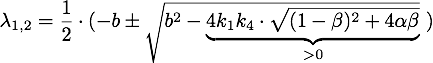
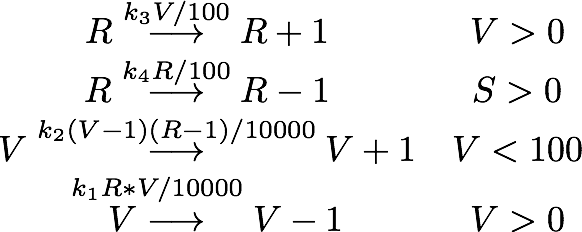
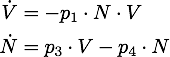
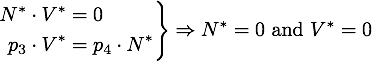
AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351