Team:Paris Saclay/Notebook/August/13
From 2013.igem.org
(→2 - Electrophoresis to check the digestion of BBa_J04450 by EcoRI/PstI) |
|||
| (2 intermediate revisions not shown) | |||
| Line 93: | Line 93: | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" |[[File:Psgel31308.jpg]] | + | | style="width:350px;border:1px solid black;" |[[File:Psgel31308.jpg|500px]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 0 : 6µL DNA Ladder | * Well 0 : 6µL DNA Ladder | ||
| Line 162: | Line 162: | ||
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We | + | We obtained a fragment at the right size. |
|} | |} | ||
Latest revision as of 01:28, 5 October 2013
Notebook : August 13
Lab work
A - Aerobic/Anaerobic regulation system
Objective : characterize BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006
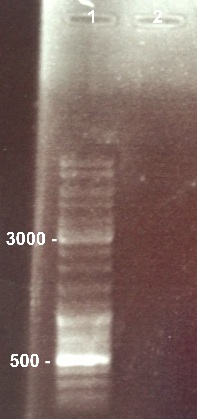
1 - Electrophoresis to check the digestion of BBa_K1155000 by SpeI/PstI
Nadia
|
|
Expected sizes :
- Pndh* : 111bp
- pSB1C3 : 2070bp
|
We can't see any band for BBa_K1155000 digestion. The digestion failed because we used the wrong enzymes. We will do it again with the good ones. |
2 - Extraction of BBa_K1155004, BBa_1155005, BBa_K1155006 from DH5αstrain
XiaoJing
Protocol : High-copy plasmid extraction
3 - Digestion of BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006 by SpeI and by SpeI/EcoRI
Anaïs, Nadia
Used quantities :
- BBa_K1155000 :
- Buffer FD : 5µL
- H2O : 39µL
- DNA : 5µL
- SpeI FD : 1µL
- BBa_K1155000 :
- Buffer FD : 5µL
- H2O : 38µL
- DNA : 5µL
- SpeI FD : 1µL
- EcoRI FD : 1µL
- BBa_K1155004, BBa_K1155005, BBa_K1155006 :
- Buffer FD : 2µL
- H2O : 7µL
- DNA : 10µL
- SpeI FD : 1µL
- BBa_K1155004, BBa_K1155005, BBa_K1155006 :
- Buffer FD : 2µL
- H2O : 6µL
- DNA : 10µL
- SpeI FD : 1µL
- EcoRI FD : 1µL
We incubat the digestion at 37°C during 10 minutes.
4 - Electrophoresis to check the digestion of BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006 by SpeI and by SpeI/EcoRI
Anaïs, Nadia
Expected sizes :
- Pndh* : 111bp
- pSB1C3 : 2070 kb
|
We lost all our digestion product so we will do it again. |
Expected sizes :
- NarK, Nar G, NirB : 200kb
- pSB1C3 : 2070bp
- NarK in pSB1C3, NarG in pSB1C3, NirB in pSB1C3 : 2270bp
|
We lost all our double digestion product so we will do it again. We obtain BBa_K1155004, BBa_K1155005, BBa_K1155006 digested by SpeI fragments at the right size. We can purify it. |
5 - Liquid culture of MG1655Z1 Δfnr::Km
XiaoJing
We picked up one colony in 5mL of LB and 5µL of kanamycine. We do it twice.
We incubated our culture at 37°C with agigation at 150rpm.
|
We obtain fragments at the right size. |
Objective : obtaining Pfnr, NarK, NarG or NirB and RBS-LacZ-Term or RBS-AmilCP-Term in pSB3K3
1 - Digestion of BBa_J04450 by EcoRI/PstI
Anaïs
Used quantities :
- Buffer FD: 2µL
- H2O : 6µL
- DNA : 10µL
- EcoRI FD : 1µL
- PstI FD : 1µL
We incubated the digestion at 37°C during 10 minutes.
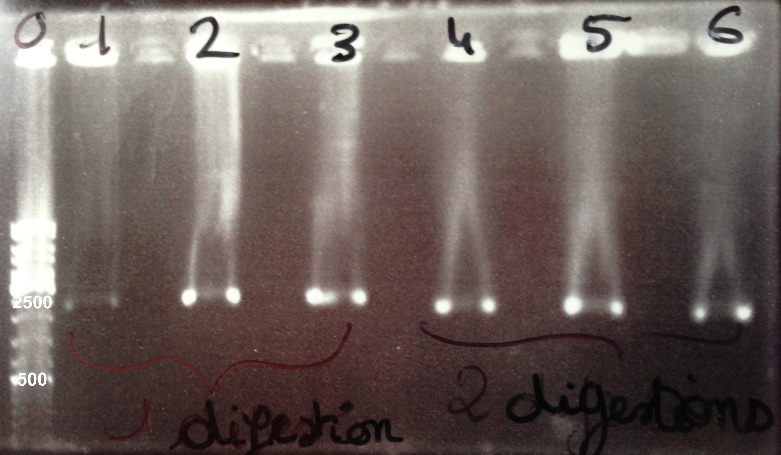
2 - Electrophoresis to check the digestion of BBa_J04450 by EcoRI/PstI
Anaïs, Nadia

|
|
Expected sizes :
- pSB3K3 : 2750bp
- GFP : 1069bp
|
We obtained a fragment at the right size. |
A - Aerobic/Anaerobic regulation system / B - PCB sensor system
Objective : obtaining FNR and BphR2 proteins (Gibson assembly)
1 - PCR of FRN Part I, FNR Part II, BphR2 Part I
Anaïs
Used quantities :
- Bphr2 Part I :
- Oligo 54F : 1µL
- Oligo 55R : 1µL
- Buffer Phusion : 10µL
- DNA of Pseudomonas pseudoalcaligenes : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
- FNR Part I :
- Oligo 59F : 1µL
- Oligo 60R : 1µL
- Buffer Phusion : 10µL
- DNA Escherichia coli : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
- FNR Part II :
- Oligo 61F : 1µL
- Oligo 62R : 1µL
- Buffer Phusion : 10µL
- DNA Escherichia coli : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
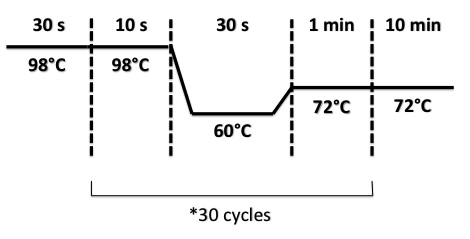
PCR Program :
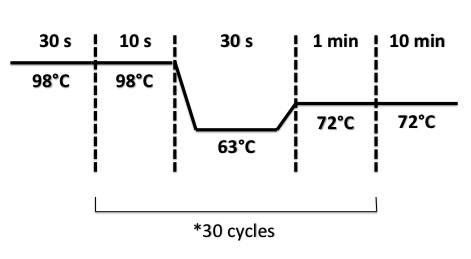
- BphR2 Part I, BphR2 Part II, RBS-BphR2 Part I :
- FNR Part I, FNR Part II, RBS-FNR Part I :
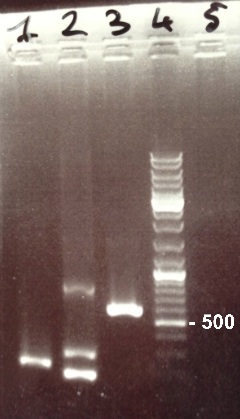
2 - Electrophoresis of PCR products : FRN Part I, FNR Part II, BphR2 Part I
Damir

|
|
|
|
Expected sizes :
- FNR Part I : 597 kb
- FNR Part II : 200 kb
- BphR2 Part I : 178 kb
|
It's impossible to read the first gel. We do it again. In the second gel, we obtain FNR Part I and FNR Part II fragments at the right size. We can purify it. We also obtain BphR2 Part I frangment at the right size but it was mix with other DNA frangments. We will try to make a gel purification of it. |
3 - Electrophoresis of PCR product : BphR2 Part I
Anaïs, Damir, Nadia

|
|
Protocol : [http://www.mn-net.com/tabid/1452/default.aspx Gel purification ]
|
Even if the two lightest stripes are overlapping, aims to do a second gel purification with these two stripes, we did the gel purification . After argumentation, we decided to do the PCR of BphR2 Part I again thanks to new PCR program and new quantities. |
4 - PCR of BphR2 Part I
Anaïs
Used quantities :
- Oligo 54F : 1µL
- Oligo 55R : 1µL
- DNA : 2µL
- Buffer Phusion : 10µL
- dNTP : 1µL
- Phusion : 1µL
- H2O : 33.5µL
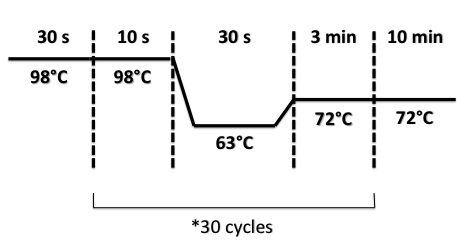
PCR Program :
5 - Gel purification of PCR product : FNR Part I, FNR Part II, RBS-FNR Part I, BphR2 Part II, RBS-BphR2 Part I'
XiaoJing
Protocol : [http://www.mn-net.com/tabid/1452/default.aspx Gel purification ]
Nanodrop :
- FNR Part I : 152.7ng/µL
- FNR Part II : 137.2ng/µL
- RBS-FNR Part I : 153.7ng/µL
- BphR2 Part II : 129.9ng/µL
- RBS-BphR2 Part I : 158.1ng/µL
|
The purification was good. We will do the Gibson assembly. |
| Previous day | Back to calendar | Next day |
 "
"