Team:Heidelberg/Templates/Indigoidine week14
From 2013.igem.org
(Created page with " ===Konrad=== ====validation of ccdB constructs==== * MP of pKH2(ccdB) constructs from independent colonies: 3rd and 4th were used according to colony PCR result ** (elution in ...") |
|||
| (3 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
===Konrad=== | ===Konrad=== | ||
====validation of ccdB constructs==== | ====validation of ccdB constructs==== | ||
| Line 192: | Line 190: | ||
QiaEXII gel extraction kit. | QiaEXII gel extraction kit. | ||
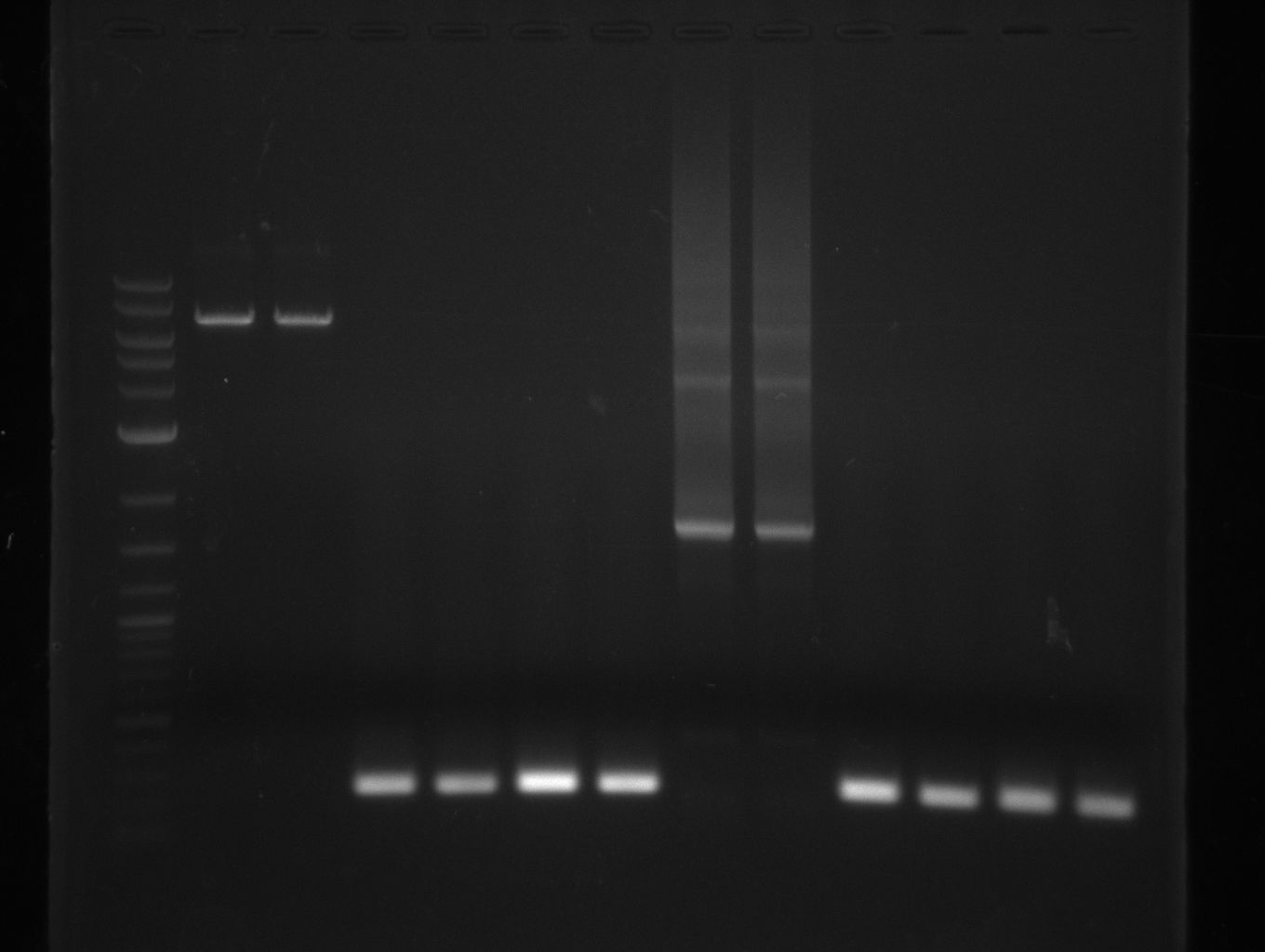
| - | + | [[File:Heidelberg_20130731_Texchainsmasscr1.jpg|1 ug 2-log; KH3/4; KH5/6; KH7/8; NI15/16; NI17/18; NI19/20]] | |
| - | + | [[File:Heidelberg_20130731_Texchainsmasscr1_cut.jpg|1 ug 2-log; KH3/4; KH5/6; KH7/8; NI15/16; NI17/18; NI19/20]] | |
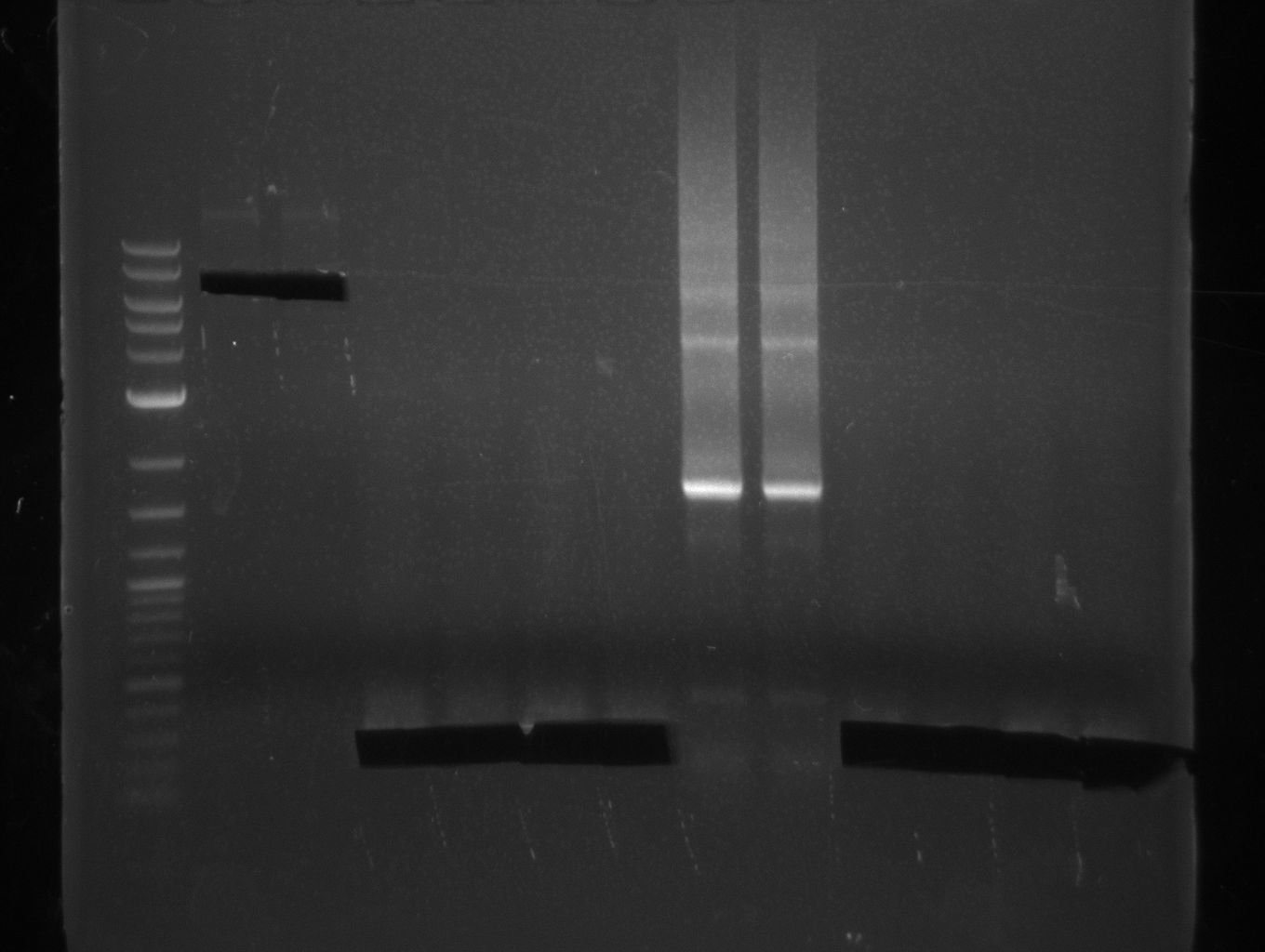
| - | File: | + | [[File:Heidelberg_20130801_Texchainsmasscr2.jpg|1 ug 2-log; NI15/16]] |
| - | File: | + | |
| - | File: | + | |
| - | + | ||
| - | + | ||
| Line 212: | Line 206: | ||
{|class="wikitable" | {|class="wikitable" | ||
|- | |- | ||
| - | !colspan="3" | + | !colspan="3"|CPEC assembly Phusion Flash HF in biometra cycler |
|- | |- | ||
!cycles!!temp (°C)!!time (s) | !cycles!!temp (°C)!!time (s) | ||
| Line 229: | Line 223: | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
On plates with colonies are always blue colonies as well as white ones -> no information on assembly success. | On plates with colonies are always blue colonies as well as white ones -> no information on assembly success. | ||
| Line 246: | Line 232: | ||
{|class="wikitable" | {|class="wikitable" | ||
|- | |- | ||
| - | !colspan="3" | + | !colspan="3"|ccdb screening pRB12 colonies with NI13/14 in |
|- | |- | ||
!cycles!!temperature (°C)!!time (s) | !cycles!!temperature (°C)!!time (s) | ||
| Line 264: | Line 250: | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
Screening is positive -> miniprep of colonies #3 and #4 from 2 ml liquid culture. | Screening is positive -> miniprep of colonies #3 and #4 from 2 ml liquid culture. | ||
Purified plasmid pRB12 was transformed to TOP10, DH5alpha and OneShot on LB+Cm to check CCDB functionality. | Purified plasmid pRB12 was transformed to TOP10, DH5alpha and OneShot on LB+Cm to check CCDB functionality. | ||
OneShot should grow whereas plates of DH5alpha and TOP10 were expected to stay empty since ccdb causes cell death. | OneShot should grow whereas plates of DH5alpha and TOP10 were expected to stay empty since ccdb causes cell death. | ||
| - | |||
| - | |||
| - | |||
All strains grow on LB+Cm which shows that ccdb doesn't work as expected. ccdb with RBS was amplified from pDONR | All strains grow on LB+Cm which shows that ccdb doesn't work as expected. ccdb with RBS was amplified from pDONR | ||
Latest revision as of 03:58, 29 October 2013
Contents |
Konrad
validation of ccdB constructs
- MP of pKH2(ccdB) constructs from independent colonies: 3rd and 4th were used according to colony PCR result
- (elution in 30 µl): col3=170 ng/µl ; col4=145 ng/µl
- testtrafo in ccdB resistent Survival, TOP10 and DH5alpha cells (from Dominik) of constructs pKH2, pKH2(ccdB)-
col3, pKH2(ccdB)-col4
- transformation with 20 ng
- plated 30 µl of cell suspension on 3 partiated plates for each constructs
Construction of ccdb-constructs pRB11-13
pRB11, pRB12 (bpsA+svpF) and pRB13 (indC+sfp) carry the ccdb gene with RBS instead of their native T-Domain so that
they don't produce the blue pigment, survive in OneShot ccdb survival cells and die when transformed to TOP10 or
DH5alpha. If the new T-Domain is successfully inserted, transformed cells survive in TOP10 and hopefully produce
the blue pigment.
amplification of CPEC fragments
NI primer in this case are for pRB11 and pRB12; KH primer for pRB13 templates are miniprepped plasmids
- ccdb (NI13/14; KH1/2; 350 bp)
- construct pKH1/2 and pRB3 without T-Domain (NI15/16; KH3/4; 7000 bp)
- indC T-Domain as a control (NI17/18; KH5/6; 200 bp)
- bpsA T-Domain as a control (NI19/20; KH7/8; 200 bp)
| component | concentration | amount |
|---|---|---|
| Phusion Flash HF MM | 2x | 25 ul |
| Primer each | 10 uM | 5 ul |
| water | 14.7 ul | |
| template | >20 ng/ul | 0.3 ul |
| NI13/14 in T100 | ||
|---|---|---|
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 8 | 98 | 1 |
| 64 ↓ 0.5 | 5 | |
| 72 | 10 | |
| 22 | 98 | 1 |
| 65 | 5 | |
| 72 | 10 | |
| 1 | 72 | 60 |
| 1 | 12 | - |
| KH1/2 in biometra | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 8 | 98 | 1 |
| 56 ↓ 0.5 | 5 | |
| 72 | 10 | |
| 22 | 98 | 1 |
| 65 | 5 | |
| 72 | 10 | |
| 1 | 72 | 60 |
| 1 | 12 | - |
| KH3/4 in T100 | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 62 ↓ 0.5 | 5 | |
| 72 | 120 | |
| 20 | 98 | 1 |
| 57 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
| NI15/16 in T100 | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
| KH5/6 in C1000 | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 63 ↓ 0.5 | 5 | |
| 72 | 10 | |
| 30 | 98 | 1 |
| 59 | 5 | |
| 72 | 10 | |
| 1 | 72 | 30 |
| 1 | 12 | - |
| NI17/18 in C1000 | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 63 ↓ 0.5 | 5 | |
| 72 | 10 | |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 10 | |
| 1 | 72 | 30 |
| 1 | 12 | - |
| KH7/8 and NI19/20 in BioRad old one | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 10 | |
| 1 | 72 | 30 |
| 1 | 12 | - |
Agarose Gel and Gel Extraction
For NI15/16 wrong template (pMM64) was used in the first PCR. The second run (pKH1/2) yielded the right product fragments have been gel extracted using QIAquick Gel extraction kit. NI15/16 and KH3/4 have been extracted using
CPEC assembly and Transformation
CPEC assembly of CCDB fragment (KH1/2 and NI13/14) and indigoidine synthesis construct (pRB3-delta-T, pKH1-delta-T
and pKH2-delta-T to yield pRB11, pRB12 and pRB13. As a positive control, native T-Domains (KH5/6 and NI19/20) are
reinserted. Transformation in ccdb resistant strain (OneShot CCDB survival cells) using standard protocol and 5 ul
of CPEC reaction product.
| CPEC assembly Phusion Flash HF in biometra cycler | ||
|---|---|---|
| cycles | temp (°C) | time (s) |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 55 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
On plates with colonies are always blue colonies as well as white ones -> no information on assembly success.
CCDB screening and test
Five white colonies from pRB12 were picked as well as a blue colony as a control. Conlony PCR using intron iTaq
polymerase was performed and 6x 0.5 mL LB+Cm were inoculated with the six colonies.
| ccdb screening pRB12 colonies with NI13/14 in | ||
|---|---|---|
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 35 | 98 | 60 |
| 60 | 30 | |
| 72 | 80 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
Screening is positive -> miniprep of colonies #3 and #4 from 2 ml liquid culture. Purified plasmid pRB12 was transformed to TOP10, DH5alpha and OneShot on LB+Cm to check CCDB functionality. OneShot should grow whereas plates of DH5alpha and TOP10 were expected to stay empty since ccdb causes cell death.
All strains grow on LB+Cm which shows that ccdb doesn't work as expected. ccdb with RBS was amplified from pDONR
plasmid.
Results and Discussion
Assembly and/or Transformation were inefficient, blue colonies on each plate suggest that a lot of template was in
the Transformation mix. The positive control doesn't provide information on whether the assembly was successful.
CCDB screenig of pRB12 was positive but transformation in DH5alpha, TOP10 and OneShot ccdb survival cells showed
that ccdb is unfunctional. In the following, all the insert from pDONR will be used including a promoter and the
ccda gene. We will work with indC on pSB1C3 with PPTases on separate plasmids to be able to easily test different
combinations of T-Domain and PPTase.
 "
"