Team:Heidelberg/Templates/Indigoidine week20
From 2013.igem.org
(Created page with " ===pRB23-T variants=== We amplified pRB23ΔccdB with KH3/4 in 2x 50 ul Phusion HF reactions and did a gel extraction. <gallery widths="400px" heights="300px"> file:201309 KH3/...") |
(→pRB23-T variants) |
||
| (3 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
===pRB23-T variants=== | ===pRB23-T variants=== | ||
We amplified pRB23ΔccdB with KH3/4 in 2x 50 ul Phusion HF reactions and did a gel extraction. | We amplified pRB23ΔccdB with KH3/4 in 2x 50 ul Phusion HF reactions and did a gel extraction. | ||
| - | |||
| - | |||
| - | |||
We performed CPEC assembly using 2.7 ul of the KH3/4 fragment and 0.3 ul of each of the T-Domain PCR products, which have been amplified some weeks ago. ''E. coli'' TOP10 were transformed with 5 ul of the CPEC reaction mix. The transformants were incubated at 37 °C over night. We picked colonies to inoculate liquid cultures for the preparation of plasmid DNA. | We performed CPEC assembly using 2.7 ul of the KH3/4 fragment and 0.3 ul of each of the T-Domain PCR products, which have been amplified some weeks ago. ''E. coli'' TOP10 were transformed with 5 ul of the CPEC reaction mix. The transformants were incubated at 37 °C over night. We picked colonies to inoculate liquid cultures for the preparation of plasmid DNA. | ||
The prepped plasmid were then used for co-transformed with each of the PPTase plasmids [[Plasmids#pRB15|pRB15-18]] as well as without a PPTase. The following table shows the transformants after four days at room temperature in the drawer. | The prepped plasmid were then used for co-transformed with each of the PPTase plasmids [[Plasmids#pRB15|pRB15-18]] as well as without a PPTase. The following table shows the transformants after four days at room temperature in the drawer. | ||
| - | + | <html> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/7/75/Slider_plattentabelle.png" alt="Co-transformation of indC-T1 to -T16 and -TTE1 to -TTE3 with and without PPTases"><br/> | |
| - | + | </html> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
synT1, synT3 and synT4 result in blue colonies!!! We sequenced the prepped plasmids using the VR. They are all correct. | synT1, synT3 and synT4 result in blue colonies!!! We sequenced the prepped plasmids using the VR. They are all correct. | ||
| Line 61: | Line 13: | ||
Transformation of TOP10 cells with the new assembled pRB16.3 variant. | Transformation of TOP10 cells with the new assembled pRB16.3 variant. | ||
| - | |||
| - | |||
| - | |||
Colony screening of six clones with VF2 and the svp reverse primer showed following result: | Colony screening of six clones with VF2 and the svp reverse primer showed following result: | ||
| - | |||
| - | |||
| - | |||
We prepped colony "3" and performed the first bulk of cotransformations with it. | We prepped colony "3" and performed the first bulk of cotransformations with it. | ||
Sequencing of the PPTase plasmids we used for the cotransformations were positive except that we found a point mutation in pRB16.3 which results in a stop codon right in the middle of svp. Therefore we will prepare the five other colonies except number 2 and screen 21 more colonies. | Sequencing of the PPTase plasmids we used for the cotransformations were positive except that we found a point mutation in pRB16.3 which results in a stop codon right in the middle of svp. Therefore we will prepare the five other colonies except number 2 and screen 21 more colonies. | ||
| - | |||
| - | |||
| - | |||
We prepped colonies 19 and 26 and cotransformed TOP10 cells together with pRB22. The shape of the colonies will show us whether the PPTase is correct or carries the stop codon. | We prepped colonies 19 and 26 and cotransformed TOP10 cells together with pRB22. The shape of the colonies will show us whether the PPTase is correct or carries the stop codon. | ||
| Line 95: | Line 38: | ||
By this we get an impression how we can detect and quantify both the total indigoidine production per well and the indigoidine production in relation to the cell density. | By this we get an impression how we can detect and quantify both the total indigoidine production per well and the indigoidine production in relation to the cell density. | ||
The measurement properties can be seen in the following figure. | The measurement properties can be seen in the following figure. | ||
| - | [[ | + | [[File:Heidelberg_Test_1_parameter.png|800px|Tecan configuration for the first test run]] |
Latest revision as of 03:53, 29 October 2013
Contents |
pRB23-T variants
We amplified pRB23ΔccdB with KH3/4 in 2x 50 ul Phusion HF reactions and did a gel extraction.
We performed CPEC assembly using 2.7 ul of the KH3/4 fragment and 0.3 ul of each of the T-Domain PCR products, which have been amplified some weeks ago. E. coli TOP10 were transformed with 5 ul of the CPEC reaction mix. The transformants were incubated at 37 °C over night. We picked colonies to inoculate liquid cultures for the preparation of plasmid DNA.
The prepped plasmid were then used for co-transformed with each of the PPTase plasmids pRB15-18 as well as without a PPTase. The following table shows the transformants after four days at room temperature in the drawer.

synT1, synT3 and synT4 result in blue colonies!!! We sequenced the prepped plasmids using the VR. They are all correct.
pRB15-19
Transformation of TOP10 cells with the new assembled pRB16.3 variant.
Colony screening of six clones with VF2 and the svp reverse primer showed following result:
We prepped colony "3" and performed the first bulk of cotransformations with it.
Sequencing of the PPTase plasmids we used for the cotransformations were positive except that we found a point mutation in pRB16.3 which results in a stop codon right in the middle of svp. Therefore we will prepare the five other colonies except number 2 and screen 21 more colonies.
We prepped colonies 19 and 26 and cotransformed TOP10 cells together with pRB22. The shape of the colonies will show us whether the PPTase is correct or carries the stop codon.
Quantitative Analysis of Indigoidine production - Assay developpement
Find best ODs to measure
We perform spectral analysis of an E. coli TOP10+pRB22+pRB15 over night liquid culture in an Amersham Biosciences Ultrospec 3300 pro. Pure TOP10 liquid culture is used as a reference and pure LB medium for blanking. The absorption spectrum hsows a significant maximum at about 590 nm.
Upscaling
To be able to increase throughput we will use a Tecan infinite M200 plate reader for our growth kinetics and indigoidine production measurement. In a first test run we measure the OD at the wavelengths 400 nm, 530 nm, 590 nm and 800 nm every 5 minutes. The liquid cultures in this case are:
- TOP10 negative control
- TOP10 + pRB22
- TOP10 + pRB3
- MG1655 + pRB3
By this we get an impression how we can detect and quantify both the total indigoidine production per well and the indigoidine production in relation to the cell density.
The measurement properties can be seen in the following figure.
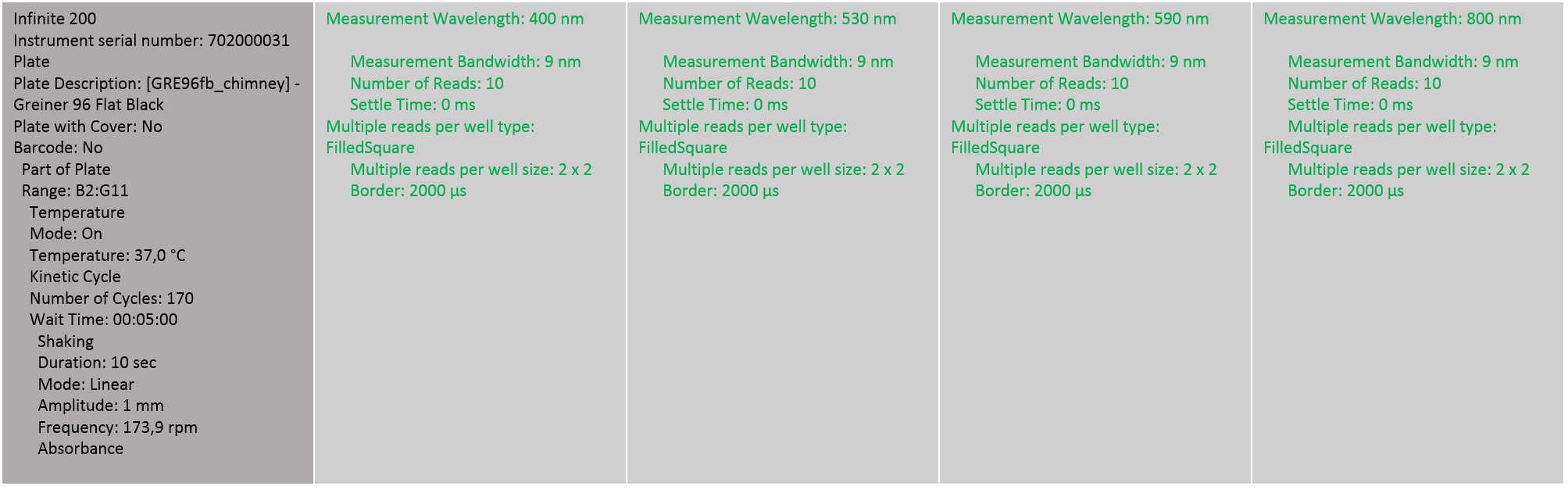
 "
"