Team:UChicago/Project
From 2013.igem.org
(→Project Overview) |
|||
| (8 intermediate revisions not shown) | |||
| Line 28: | Line 28: | ||
== '''Project Overview''' == | == '''Project Overview''' == | ||
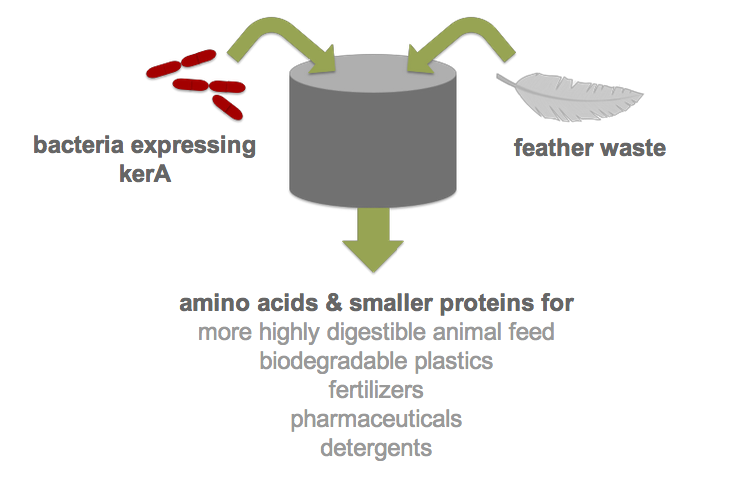
| - | Each year, over two billion pounds of feather waste is produced as a byproduct of commercial poultry processing. Most of this waste is chemically treated or steam pressure-cooked into nutrient-poor animal feed (1). Nonetheless, recent research has shown that feather keratin can be used to produce biodegradable plastics (2), fertilizers (3), detergents (4), and pharmaceuticals (5). However, traditional methods of keratin degradation rely on chemicals such as sulfides and limes which are energetically costly and pose serious environmental and health concerns. Therefore, researchers have turned to feather-degrading bacteria as an alternative to traditional chemical methods (6). Although keratin-degrading bacterial strains were isolated from poultry waste nearly two decades ago and some keratin-degrading proteases (keratinases) have been identified, keratinase production in heterologous hosts has been stymied by poor protease expression. | + | Each year, over two billion pounds of feather waste is produced as a byproduct of commercial poultry processing. Most of this waste is chemically treated or steam pressure-cooked into nutrient-poor animal feed (1). Nonetheless, recent research has shown that feather keratin can be used to produce biodegradable plastics (2), fertilizers (3), detergents (4), and pharmaceuticals (5). However, traditional methods of keratin degradation rely on chemicals such as sulfides and limes which are energetically costly and pose serious environmental and health concerns. Therefore, researchers have turned to feather-degrading bacteria as an alternative to traditional chemical methods (6). Although keratin-degrading bacterial strains were isolated from poultry waste nearly two decades ago and some keratin-degrading proteases (keratinases) have been identified, keratinase production in heterologous hosts has been stymied by poor protease expression. By developing an efficient process for the digestion of keratin, we would not only drastically reduce the cost of disposal but also allow the keratin to be used in products like animal feed, fertilizers and biodegradable plastics. Our final product would be a one-step continuous process in which feather waste could be continually added to <i>B. subtilis</i> cultures. |
| + | |||
| + | |||
| + | [[File:Feather_degradation_schematic.jpg|frame|A simple schematic of our project]] | ||
| + | |||
| + | |||
| Line 38: | Line 43: | ||
We also wanted to determine if KerA expression in E. coli BL21-DE3 was comparable to that of B. subtilis. Unlike the members of the Bacillus genus, E. coli has two membranes. The kerA signal peptide could allow transport across the inner membrane but not the outer membrane, trapping KerA in the periplasm, where the amino acid modifications could render the keratinase inactive. Moreover, it is possible that E. coli does not contain the cellular machinery necessary to cleave the kerA signal peptide, producing an inactive form of the enzyme. Thus, we designed an alternate kerA BioBrick that was missing the signal peptide. To optimize expression of the keratinase in E. coli, we would have used a constitutive E. coli promoter BioBrick and a high copy number backbone. | We also wanted to determine if KerA expression in E. coli BL21-DE3 was comparable to that of B. subtilis. Unlike the members of the Bacillus genus, E. coli has two membranes. The kerA signal peptide could allow transport across the inner membrane but not the outer membrane, trapping KerA in the periplasm, where the amino acid modifications could render the keratinase inactive. Moreover, it is possible that E. coli does not contain the cellular machinery necessary to cleave the kerA signal peptide, producing an inactive form of the enzyme. Thus, we designed an alternate kerA BioBrick that was missing the signal peptide. To optimize expression of the keratinase in E. coli, we would have used a constitutive E. coli promoter BioBrick and a high copy number backbone. | ||
| - | |||
| - | |||
== Sources == | == Sources == | ||
Latest revision as of 02:54, 26 October 2013
Project Overview
Each year, over two billion pounds of feather waste is produced as a byproduct of commercial poultry processing. Most of this waste is chemically treated or steam pressure-cooked into nutrient-poor animal feed (1). Nonetheless, recent research has shown that feather keratin can be used to produce biodegradable plastics (2), fertilizers (3), detergents (4), and pharmaceuticals (5). However, traditional methods of keratin degradation rely on chemicals such as sulfides and limes which are energetically costly and pose serious environmental and health concerns. Therefore, researchers have turned to feather-degrading bacteria as an alternative to traditional chemical methods (6). Although keratin-degrading bacterial strains were isolated from poultry waste nearly two decades ago and some keratin-degrading proteases (keratinases) have been identified, keratinase production in heterologous hosts has been stymied by poor protease expression. By developing an efficient process for the digestion of keratin, we would not only drastically reduce the cost of disposal but also allow the keratin to be used in products like animal feed, fertilizers and biodegradable plastics. Our final product would be a one-step continuous process in which feather waste could be continually added to B. subtilis cultures.
In order to improve keratinase expression in Bacillus subtilis, a host suited to industrial use, we decided to build a BioBrick for the best characterized and most efficient keratinase gene. The keratin-degrading bacterial strain that is best characterized in the literature is Bacillus licheniformis PWD-1. The keratinase, called KerA, responsible for this bacterium’s activity was isolated and characterized by Shih et al. along with its signal peptide and promoter (7). The DNA sequence of kerA is 1458 bases long and codes for a serine protease. Like many secreted proteases in the Bacillus genus, KerA has a signal peptide, which is cleaved before KerA crosses the single plasma membrane of B. licheniformis. Additionally, the kerA promoter is activated when keratin is the sole source of carbon and nitrogen. Shih et al. successfully transformed B. subtilis with kerA and expressed it with its own promoter as well as with P43, a constitutive promoter, and a combination of the two in a high copy number backbone. All transformants expressed keratinase activity in the presence and absence of keratin (8).
Building on the success of Shih et al., our team designed a BioBrick to improve kerA expression in a heterologous host. We purchased two gBlocks from IDT to assemble the kerA DNA sequence, which included the signal peptide. In order to increase the level of KerA produced in B. subtilis, we planned to use BioBrick part BBa_K143053, which includes Pveg, a constitutive promoter. Moreover, a high copy number backbone with an origin of replication for B. subtilis is needed for the largest production of KerA. Since the current B. subtilis plasmids in the iGEM registry are single copy plasmids that integrate into the genome, our team designed a high copy number backbone compatible with B. subtilis based on pUB110 (9).
We also wanted to determine if KerA expression in E. coli BL21-DE3 was comparable to that of B. subtilis. Unlike the members of the Bacillus genus, E. coli has two membranes. The kerA signal peptide could allow transport across the inner membrane but not the outer membrane, trapping KerA in the periplasm, where the amino acid modifications could render the keratinase inactive. Moreover, it is possible that E. coli does not contain the cellular machinery necessary to cleave the kerA signal peptide, producing an inactive form of the enzyme. Thus, we designed an alternate kerA BioBrick that was missing the signal peptide. To optimize expression of the keratinase in E. coli, we would have used a constitutive E. coli promoter BioBrick and a high copy number backbone.
Sources
(1) El-Nagar, K., Saleh, S. M., & Ramadan, A. R. (2006). [http://www.tx.ncsu.edu/jtatm/volume5issue2/articles/El-Nagar/El_nagar_Full_171-05.pdf Utilization of feather waste to improve the properties of the Egyptian cotton fabrics.] Jour. of Textile and Apparel Technology and Management, 2, 1-12.
(2) Jin, E., Reddy, N., Zhu, Z., & Yang, Y. (2011). [http://pubs.acs.org/doi/abs/10.1021/jf1039519 Graft polymerization of native chicken feathers for thermoplastic applications.] Journal of agricultural and food chemistry, 59(5), 1729-1738.
(3) Hadas, A., & Kautsky, L. (1994). [http://link.springer.com/article/10.1007%2FBF00748776 Feather meal, a semi-slow-release nitrogen fertilizer for organic farming.] Fertilizer research, 38(2), 165-170.
(4) Riffel, A., Lucas, F., Heeb, P., & Brandelli, A. (2003). [http://www.ncbi.nlm.nih.gov/pubmed/12677362 Characterization of a new keratinolytic bacterium that completely degrades native feather keratin.] Archives of Microbiology, 179(4), 258-265.
(5) Tiwary, E., & Gupta, R. (2010). [http://www.ncbi.nlm.nih.gov/pubmed/20597544 Extracellular expression of keratinase from Bacillus licheniformis ER-15 in Escherichia coli.] Journal of agricultural and food chemistry, 58(14), 8380-8385.
(6) Williams, C. M., Richter, C. S., Mackenzie, J. M., & Shih, J. C. (1990). [http://aem.asm.org/content/56/6/1509 Isolation, identification, and characterization of a feather-degrading bacterium.] Applied and Environmental Microbiology, 56(6), 1509-1515.
(7) Lin, X., Kelemen, D. W., Miller, E. S., & Shih, J. C. (1995). [http://www.ncbi.nlm.nih.gov/pubmed/7747965 Nucleotide sequence and expression of kerA, the gene encoding a keratinolytic protease of Bacillus licheniformis PWD-1.] Applied and environmental microbiology,61(4), 1469-1474.
(8) Lin, X., Wong, S. L., Miller, E. S., & Shih, J. C. H. (1997). [http://www.ncbi.nlm.nih.gov/pubmed/9366094 Expression of the Bacillus licheniformis PWD-1 keratinase gene in B. subtilis.] Journal of Industrial Microbiology and Biotechnology, 19(2), 134-138.
(9) Boe, L. A. R. S., Gros, M. F., te Riele, H. E. I. N., Ehrlich, S. D., & Gruss, A. (1989). [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC210059/ Replication origins of single-stranded-DNA plasmid pUB110.] Journal of bacteriology, 171(6), 3366-3372.
Results
Check out our BioBrick designs.
 "
"