Team:Stanford-Brown/Projects/EuCROPIS
From 2013.igem.org
Vecchionis (Talk | contribs) (→Accomplishments) |
|||
| (28 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<div class="center" style="width: auto; margin-left: auto; margin-right: auto;">''' '''</div> | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">''' '''</div> | ||
| + | ===Satellite Mission=== | ||
* Constructed and synthesized prototype chromogenic biosensors to detect sucrose induction and sporulation | * Constructed and synthesized prototype chromogenic biosensors to detect sucrose induction and sporulation | ||
* Contributed on project to validate PowerCell on the EuCROPIS satellite mission and send synthetic biology into outer space | * Contributed on project to validate PowerCell on the EuCROPIS satellite mission and send synthetic biology into outer space | ||
| - | Submitted 5 BioBricks | + | ===Submitted 5 BioBricks=== |
* BBa_K1218020 | * BBa_K1218020 | ||
* BBa_K1218001 | * BBa_K1218001 | ||
| Line 15: | Line 16: | ||
* BBa_K1218025 | * BBa_K1218025 | ||
| + | <br/> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| Line 27: | Line 29: | ||
In 2016, the German Aerospace Center will be launching the Euglena: Combined Regenerative Organic-Food Production In Space (EuCROPIS) satellite mission into low-earth orbit. While the mission's main objective is to study food production, the Germans have invited several other researchers, including astrobiology researchers from NASA, to participate in the study by sending their own experiments along as secondary payloads. As a result, we are proud to announce that we will be using the EuCROPIS mission as a way to test synthetic biology in space for the first time! | In 2016, the German Aerospace Center will be launching the Euglena: Combined Regenerative Organic-Food Production In Space (EuCROPIS) satellite mission into low-earth orbit. While the mission's main objective is to study food production, the Germans have invited several other researchers, including astrobiology researchers from NASA, to participate in the study by sending their own experiments along as secondary payloads. As a result, we are proud to announce that we will be using the EuCROPIS mission as a way to test synthetic biology in space for the first time! | ||
| - | In 2011, the Brown-Stanford iGEM team built PowerCell, a photosynthetic and nitrogen-fixing cyanobacterium engineered to secrete sucrose. The project addressed one of NASA's key concerns regarding the future of terraforming Mars, the need for in-situ resource utilization. Since sending materials into space is very expensive, engineering an organism that could be used as an energy source was key. Through the secretion of its naturally-generated sucrose, PowerCell addressed this need effectively by providing other bacterial cultures with a rich carbon source that can ideally be used to produce biomass such as food or drugs. | + | In 2011, the [https://2011.igem.org/Team:Brown-Stanford/PowerCell/Introduction '''Brown-Stanford iGEM team built PowerCell,'''] a photosynthetic and nitrogen-fixing cyanobacterium engineered to secrete sucrose. The project addressed one of NASA's key concerns regarding the future of terraforming Mars, the need for in-situ resource utilization. Since sending materials into space is very expensive, engineering an organism that could be used as an energy source was key. Through the secretion of its naturally-generated sucrose, PowerCell addressed this need effectively by providing other bacterial cultures with a rich carbon source that can ideally be used to produce biomass such as food or drugs. |
Despite the results generated by the 2011 team in the lab, however, the need to assess PowerCell's effectiveness in space arose, which is where we come in. | Despite the results generated by the 2011 team in the lab, however, the need to assess PowerCell's effectiveness in space arose, which is where we come in. | ||
| - | The goal of our EuCROPIS project was to find a way to assess the efficacy of PowerCell while aboard the German satellite mission. Due to the constraints provided by the mission - such as the need for the experimental organisms to survive the conditions of outer space in addition to months or years waiting on the launchpad, the fact that the satellite can only measure data via a spectrophotometer, and the experimental constraints posed by using Pharmasat microfluidic cards - we decided to validate PowerCell by building a chromogenic biosensor in <i>Bacillus subtilis</i>. | + | The goal of our EuCROPIS project was to find a way to assess the efficacy of PowerCell while aboard the German satellite mission. Due to the constraints provided by the mission - such as the need for the experimental organisms to survive the conditions of outer space in addition to months or years waiting on the launchpad, the fact that the satellite can only measure data via a spectrophotometer, and the experimental constraints posed by using Pharmasat microfluidic cards - we decided to validate PowerCell by building a chromogenic biosensor in <i>Bacillus subtilis</i> (Horneck et al., 2010). |
| - | We chose to work with the bacterium <i>B. subtilis</i> in developing our biosensor because it is a hardy organism that forms endospores in nutrient-depleted conditions and can survive for years in the environmental extremes found in space (cold, desiccation, pressure, low gravity, etc.). When it is exposed to nutrients, however, such as sucrose, the bacterium undergoes germination. As a result, we' | + | We chose to work with the bacterium <i>B. subtilis</i> in developing our biosensor because it is a hardy organism that forms endospores in nutrient-depleted conditions and can survive for years in the environmental extremes found in space (cold, desiccation, pressure, low gravity, etc.). When it is exposed to nutrients, however, such as sucrose, the bacterium undergoes germination (Horneck, 1992). As a result, we're able to characterize promoters associated with sucrose induction (sacY) and sporulation (spo0A) with chromoproteins in varying colors developed by the [https://2012.igem.org/Team:Uppsala_University/Chromoproteins '''Uppsala iGEM team'''] in 2012 (Crutz et al., 1992). By linking these colorimetric proteins to our genes of interest, we will be able to monitor the state our biosensor is in and see whether it is able to make use of the sucrose being provided by the PowerCell that it is cultured with within the wells of the microfluidics card. The color changes associated with the change in states can be picked up by the spectrophotometer and reported back to us here on earth, thereby letting us know how the first synthetically biological organism to be sent up into space is faring! |
== '''Construct Assembly''' == | == '''Construct Assembly''' == | ||
| - | We began by researching and identifying genes associated with sporulation, germination, and sucrose induction in <i>Bacillus subtilis</i>. We decided to focus on sacY, a gene the regulates sucrose induction and is itself induced by the presence of sucrose, and spo0A, a gene that regulates sporulation by influencing more than 500 genes related to sporulation. We attempted to isolate the genes from the genome using PCR with mixed results. We decided instead to construct the two promoters by annealing complimentary oligos from Elim, and using PCR to add the BioBrick ends. | + | We began by researching and identifying genes associated with sporulation, germination, and sucrose induction in <i>Bacillus subtilis</i>. We decided to focus on sacY, a gene the regulates sucrose induction and is itself induced by the presence of sucrose (Crutz and Steinmentz, 1992), and spo0A, a gene that regulates sporulation by influencing more than 500 genes related to sporulation (Chibazakura et al., 1991). We attempted to isolate the genes from the genome using PCR with mixed results. We decided instead to construct the two promoters by annealing complimentary oligos from Elim, and using PCR to add the BioBrick ends. |
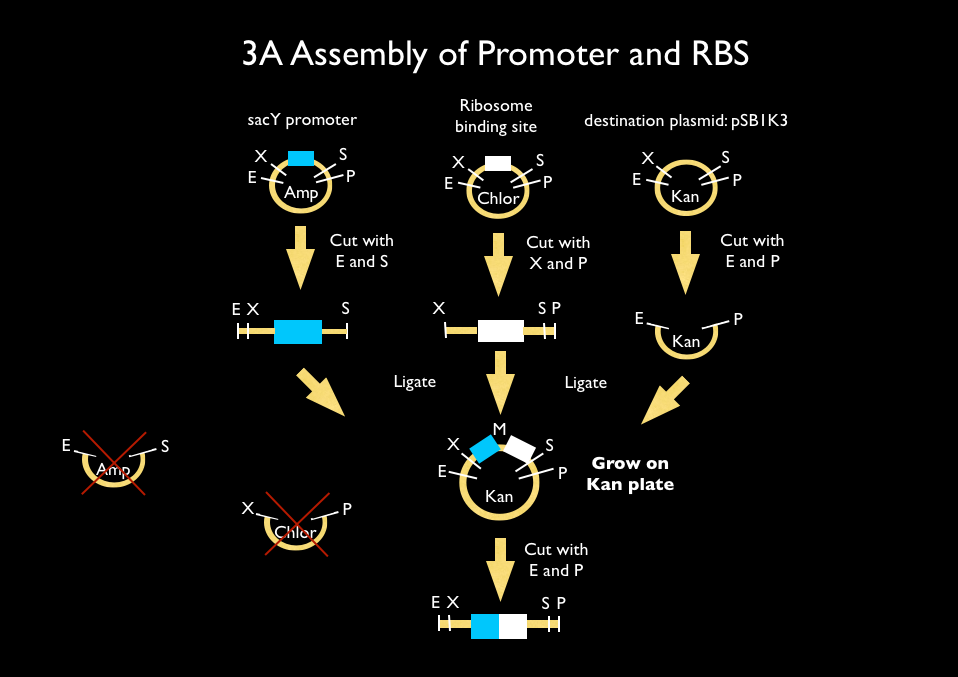
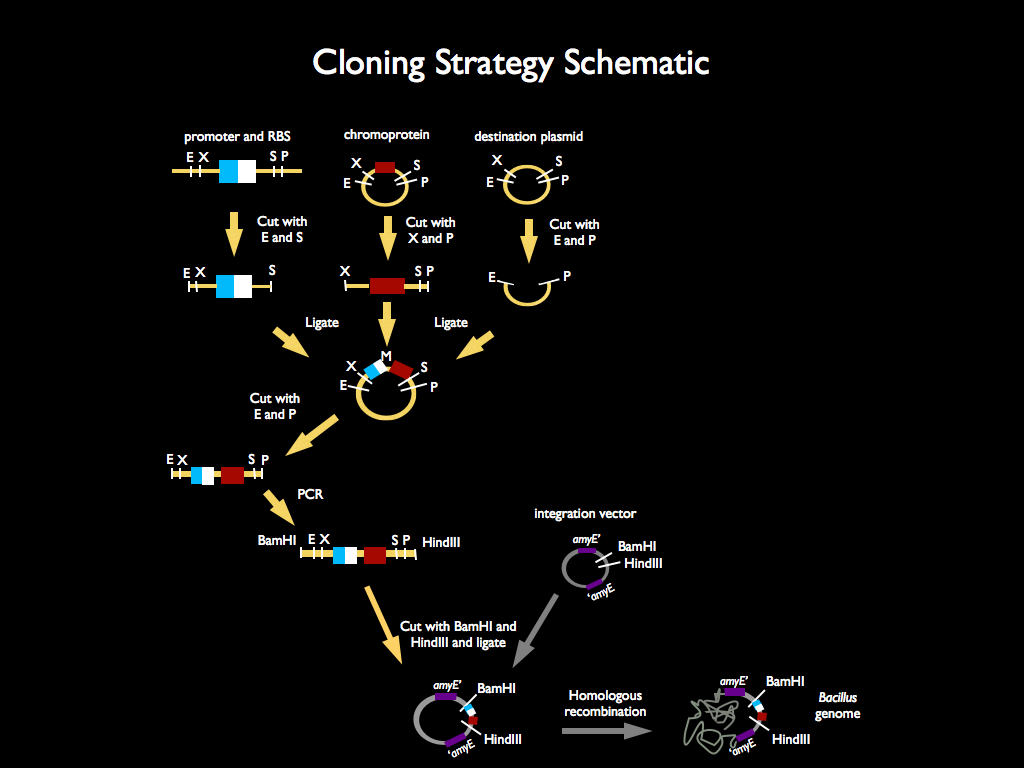
| - | We used 3A assembly to add a ribosome binding site from the registry (BBa_B0034) directly downstream of our sucrose inducer promoter (see Figure A). After an E/P digestion, we proceeded to another 3A assembly to add a red chromoprotein ( | + | We used 3A assembly to add a ribosome binding site from the registry (BBa_B0034) directly downstream of our sucrose inducer promoter (see Figure A). After an E/P digestion, we proceeded to another 3A assembly to add a red chromoprotein (BBa_K592012) downstream of the sucrose inducer promoter and ribosome binding site (Figure B). Likewise we did another assembly in parallel to add a red fluorescent protein (BBa_E1010) downstream of the sucrose inducer promoter and ribosome binding site. We then used primers from Elim and PCR to add the restriction sites BamHI and HindIII to either side of our two constructs. In addition, we ordered two constructs from DNA 2.0: the sucrose inducing promoter linked to eForRed and the sporulation regulator promoter linked to a blue chromoprotein (BBa_K864401). We designed the constructs such that they were flanked with the restriction sites BamHI and HindIII. These two restriction sites allowed us to ligate each of our constructs into <i>Bacillus subtilis</i> integration vectors (BGSC: ECE115). Finally, we used electroporation to transform into electrocompetent <i>Bacillus subtilis</i> cells. |
[[File:FigureA.jpg|center|750px]] | [[File:FigureA.jpg|center|750px]] | ||
| Line 53: | Line 55: | ||
The primary protocols for the EuCROPIS project can be found [https://docs.google.com/document/d/1LjKxN-iKf7vPodXDk3tc8yI7lC7L5JQPntps6gGhAgM/edit# '''here''']. | The primary protocols for the EuCROPIS project can be found [https://docs.google.com/document/d/1LjKxN-iKf7vPodXDk3tc8yI7lC7L5JQPntps6gGhAgM/edit# '''here''']. | ||
| - | Our team has compiled a full list or our protocols and lab techniques [https://docs.google.com/a/ | + | Our team has compiled a full list or our protocols and lab techniques [https://docs.google.com/a/brown.edu/document/d/10hpfdvthGIWRPrrQCnWNIy3HfinFoK0hlV7VMb5nGHI '''here''']. |
== '''Lab Notebook''' == | == '''Lab Notebook''' == | ||
| Line 61: | Line 63: | ||
== '''Data''' == | == '''Data''' == | ||
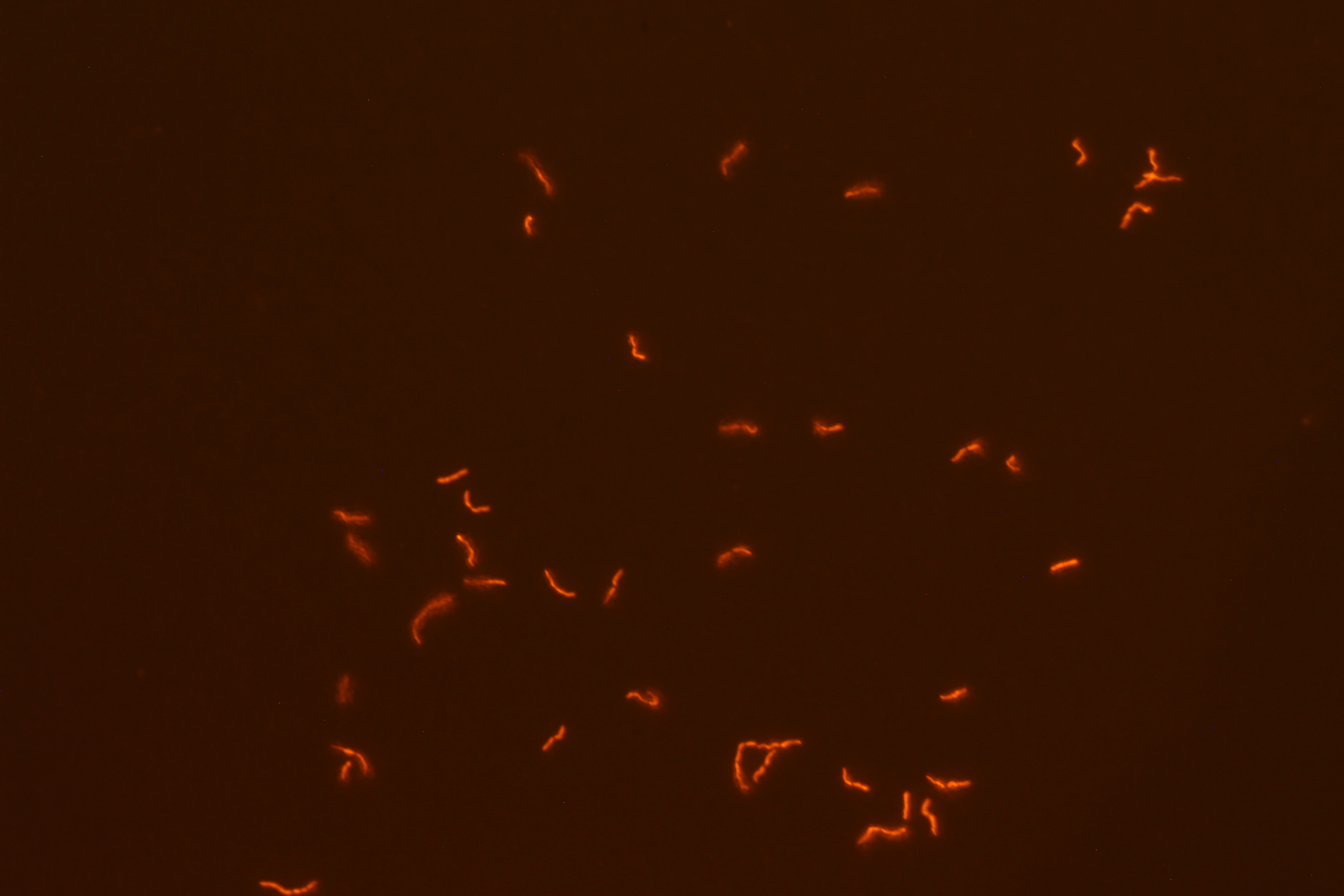
| - | We successfully built constructs linking our two promoters of interest with chromoproteins and fluorescent proteins of varying colors. Using an integration vector, we transformed our constructs into <i>B. subtilis</i>. Microscopy reveals that some of our <i>B. subtilis</i> cells were transformed successfully, specifically for the construct that links the sucrose inducer promoter with a red chromoprotein that has fluorescent properties (Figure 4) and the construct that links the sucrose inducer promoter with a red fluorescent protein (Figure 6). The controls which contain the integration vector but no construct did not fluoresce. | + | We successfully built constructs linking our two promoters of interest with chromoproteins and fluorescent proteins of varying colors. Using an integration vector, we transformed our constructs into <i>B. subtilis</i>. Microscopy reveals that some of our <i>B. subtilis</i> cells were transformed successfully, specifically for the construct that links the sucrose inducer promoter with a red chromoprotein that has fluorescent properties (Figure 4) and the construct that links the sucrose inducer promoter with a red fluorescent protein (Figure 6). The controls which contain the integration vector but no construct did not fluoresce. Since we did not add additional sucrose to the medium, it is possible the fluorescence from our engineered B. subtilis is due to promoter leakiness, trace sucrose in the growth medium, or an unknown biological contaminant. We are currently performing experiments to test these explanations and to further characterize our promoters. We are currently performing experiments to test these explanations and to further characterize our promoters. |
First, we are testing minimal media solutions to prove that the <i>B. subtilis</i> does not fluoresce except in the presence of sucrose. Second, comparing Figures 3 and 4 illustrates that many of the cells are not <i>B. subtilis</i> cells containing our constructs. This is in part due to an unknown contaminant. The contaminant and the unusual lawn-like morphology of our <i>B. subtilis</i> transformant when grown on a plate make it impossible for us to find and pick individual colonies containing the proper construct. We are in the process of solving this problem, and when we do, we will be able to qualitatively and quantitatively test color change and fluorescence in response to sucrose and sporulation medium. | First, we are testing minimal media solutions to prove that the <i>B. subtilis</i> does not fluoresce except in the presence of sucrose. Second, comparing Figures 3 and 4 illustrates that many of the cells are not <i>B. subtilis</i> cells containing our constructs. This is in part due to an unknown contaminant. The contaminant and the unusual lawn-like morphology of our <i>B. subtilis</i> transformant when grown on a plate make it impossible for us to find and pick individual colonies containing the proper construct. We are in the process of solving this problem, and when we do, we will be able to qualitatively and quantitatively test color change and fluorescence in response to sucrose and sporulation medium. | ||
| Line 102: | Line 104: | ||
|} | |} | ||
</center> | </center> | ||
| + | |||
| + | |||
| + | == '''Conclusions''' == | ||
| + | |||
| + | Through this project, we constructed and synthesized prototype chromogenic biosensors to detect sucrose metabolism and sporulation in <i>Bacillus subtilis</i>. Fluorescent micrographs show that our transformations were successful and that our engineered biosensors fluoresce. However, we still have further work to do in order to isolate the fluorescent colonies and characterize our promoters of interest, which may be exhibiting promoter leakiness. This future work will be important to prepare for the 2016 EuCROPIS satellite mission and a helium balloon test next year. | ||
== '''BioBricks''' == | == '''BioBricks''' == | ||
| Line 123: | Line 130: | ||
* Dr. Daniel R. Zeigler, BGSC Director and the Bacillus Genetic Stock Center — Free strains of Bacillus integration vectors | * Dr. Daniel R. Zeigler, BGSC Director and the Bacillus Genetic Stock Center — Free strains of Bacillus integration vectors | ||
* Dr. Elwood Agasid — Explanation of EuCROPIS mission from an engineering perspective | * Dr. Elwood Agasid — Explanation of EuCROPIS mission from an engineering perspective | ||
| + | |||
| + | |||
| + | == '''References''' == | ||
| + | |||
| + | *Chibazakura, T., Kawamura, F., Takahashi, H. (1991). Differential Regulation of ''spo0A'' Transcription in ''Bacillus subtilis'': Glucose Represses Promoter Switching at the Initiation of Sporulation. Journal of Bacteriology 173: 2625-2632. | ||
| + | *Crutz, A., Steinmetz, M. (1992). Transcription of the ''Bacillus subtilis sacX'' and ''sacY'' Genes, Encoding Regulators of Sucrose Metabolism, Is Both Inducible by Sucrose and Controlled by the DegS-DegU Signalling System. Journal of Bacteriology 174: 6087-6095. | ||
| + | *Hauslage, J., Lebert, M., Strauch, S. M., Mancinelli, R. Eu:CROPIS - Euglena: Combined Regenerative Organic-Food Production In Space. Space Definition Document. | ||
| + | *Horneck, G. (1992). Responses of ''Bacillus Subtilis'' spores to space environment: Results from experiments in space. Microbiology and Molecular Biology Reviews. Origins of Life and Evolution of the Biosphere 23: 37-52. | ||
| + | *Horneck, G., Klaus, D. M., Mancinelli, R. L. (2010). Space Microbiology. Microbiology and Molecular Biology Reviews 74: 121-156. | ||
| + | *Keijser, B. J. F., Beek, A. T., Rauwerda, H., Schuren, F., Montijn, R., Van der Spek, H., Brul, S. (2007). Analysis of Temporal Gene Expression during "Bacillus subtilis" Spore Germination and Outgrowth. Journal of Bacteriology 189: 3624-3634. | ||
Latest revision as of 03:02, 29 October 2013
Accomplishments
Satellite Mission
- Constructed and synthesized prototype chromogenic biosensors to detect sucrose induction and sporulation
- Contributed on project to validate PowerCell on the EuCROPIS satellite mission and send synthetic biology into outer space
Submitted 5 BioBricks
- BBa_K1218020
- BBa_K1218001
- BBa_K1218021
- BBa_K1218023
- BBa_K1218025
Introduction
In 2016, the German Aerospace Center will be launching the Euglena: Combined Regenerative Organic-Food Production In Space (EuCROPIS) satellite mission into low-earth orbit. While the mission's main objective is to study food production, the Germans have invited several other researchers, including astrobiology researchers from NASA, to participate in the study by sending their own experiments along as secondary payloads. As a result, we are proud to announce that we will be using the EuCROPIS mission as a way to test synthetic biology in space for the first time!
In 2011, the Brown-Stanford iGEM team built PowerCell, a photosynthetic and nitrogen-fixing cyanobacterium engineered to secrete sucrose. The project addressed one of NASA's key concerns regarding the future of terraforming Mars, the need for in-situ resource utilization. Since sending materials into space is very expensive, engineering an organism that could be used as an energy source was key. Through the secretion of its naturally-generated sucrose, PowerCell addressed this need effectively by providing other bacterial cultures with a rich carbon source that can ideally be used to produce biomass such as food or drugs.
Despite the results generated by the 2011 team in the lab, however, the need to assess PowerCell's effectiveness in space arose, which is where we come in.
The goal of our EuCROPIS project was to find a way to assess the efficacy of PowerCell while aboard the German satellite mission. Due to the constraints provided by the mission - such as the need for the experimental organisms to survive the conditions of outer space in addition to months or years waiting on the launchpad, the fact that the satellite can only measure data via a spectrophotometer, and the experimental constraints posed by using Pharmasat microfluidic cards - we decided to validate PowerCell by building a chromogenic biosensor in Bacillus subtilis (Horneck et al., 2010).
We chose to work with the bacterium B. subtilis in developing our biosensor because it is a hardy organism that forms endospores in nutrient-depleted conditions and can survive for years in the environmental extremes found in space (cold, desiccation, pressure, low gravity, etc.). When it is exposed to nutrients, however, such as sucrose, the bacterium undergoes germination (Horneck, 1992). As a result, we're able to characterize promoters associated with sucrose induction (sacY) and sporulation (spo0A) with chromoproteins in varying colors developed by the Uppsala iGEM team in 2012 (Crutz et al., 1992). By linking these colorimetric proteins to our genes of interest, we will be able to monitor the state our biosensor is in and see whether it is able to make use of the sucrose being provided by the PowerCell that it is cultured with within the wells of the microfluidics card. The color changes associated with the change in states can be picked up by the spectrophotometer and reported back to us here on earth, thereby letting us know how the first synthetically biological organism to be sent up into space is faring!
Construct Assembly
We began by researching and identifying genes associated with sporulation, germination, and sucrose induction in Bacillus subtilis. We decided to focus on sacY, a gene the regulates sucrose induction and is itself induced by the presence of sucrose (Crutz and Steinmentz, 1992), and spo0A, a gene that regulates sporulation by influencing more than 500 genes related to sporulation (Chibazakura et al., 1991). We attempted to isolate the genes from the genome using PCR with mixed results. We decided instead to construct the two promoters by annealing complimentary oligos from Elim, and using PCR to add the BioBrick ends.
We used 3A assembly to add a ribosome binding site from the registry (BBa_B0034) directly downstream of our sucrose inducer promoter (see Figure A). After an E/P digestion, we proceeded to another 3A assembly to add a red chromoprotein (BBa_K592012) downstream of the sucrose inducer promoter and ribosome binding site (Figure B). Likewise we did another assembly in parallel to add a red fluorescent protein (BBa_E1010) downstream of the sucrose inducer promoter and ribosome binding site. We then used primers from Elim and PCR to add the restriction sites BamHI and HindIII to either side of our two constructs. In addition, we ordered two constructs from DNA 2.0: the sucrose inducing promoter linked to eForRed and the sporulation regulator promoter linked to a blue chromoprotein (BBa_K864401). We designed the constructs such that they were flanked with the restriction sites BamHI and HindIII. These two restriction sites allowed us to ligate each of our constructs into Bacillus subtilis integration vectors (BGSC: ECE115). Finally, we used electroporation to transform into electrocompetent Bacillus subtilis cells.
Protocols
The primary protocols for the EuCROPIS project can be found here.
Our team has compiled a full list or our protocols and lab techniques here.
Lab Notebook
The EuCROPIS lab notebook can be accessed here.
Data
We successfully built constructs linking our two promoters of interest with chromoproteins and fluorescent proteins of varying colors. Using an integration vector, we transformed our constructs into B. subtilis. Microscopy reveals that some of our B. subtilis cells were transformed successfully, specifically for the construct that links the sucrose inducer promoter with a red chromoprotein that has fluorescent properties (Figure 4) and the construct that links the sucrose inducer promoter with a red fluorescent protein (Figure 6). The controls which contain the integration vector but no construct did not fluoresce. Since we did not add additional sucrose to the medium, it is possible the fluorescence from our engineered B. subtilis is due to promoter leakiness, trace sucrose in the growth medium, or an unknown biological contaminant. We are currently performing experiments to test these explanations and to further characterize our promoters. We are currently performing experiments to test these explanations and to further characterize our promoters.
First, we are testing minimal media solutions to prove that the B. subtilis does not fluoresce except in the presence of sucrose. Second, comparing Figures 3 and 4 illustrates that many of the cells are not B. subtilis cells containing our constructs. This is in part due to an unknown contaminant. The contaminant and the unusual lawn-like morphology of our B. subtilis transformant when grown on a plate make it impossible for us to find and pick individual colonies containing the proper construct. We are in the process of solving this problem, and when we do, we will be able to qualitatively and quantitatively test color change and fluorescence in response to sucrose and sporulation medium.
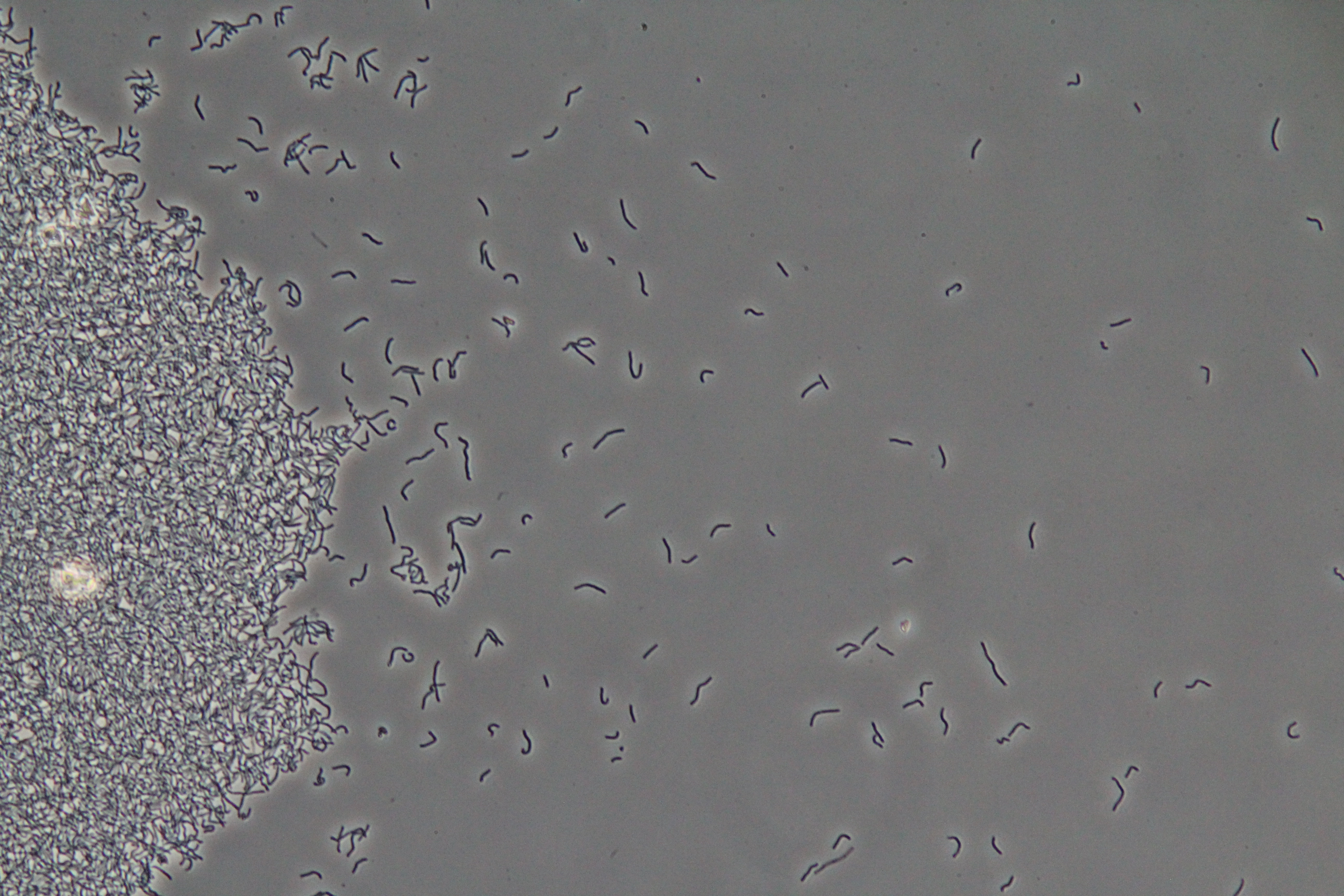
| 
|
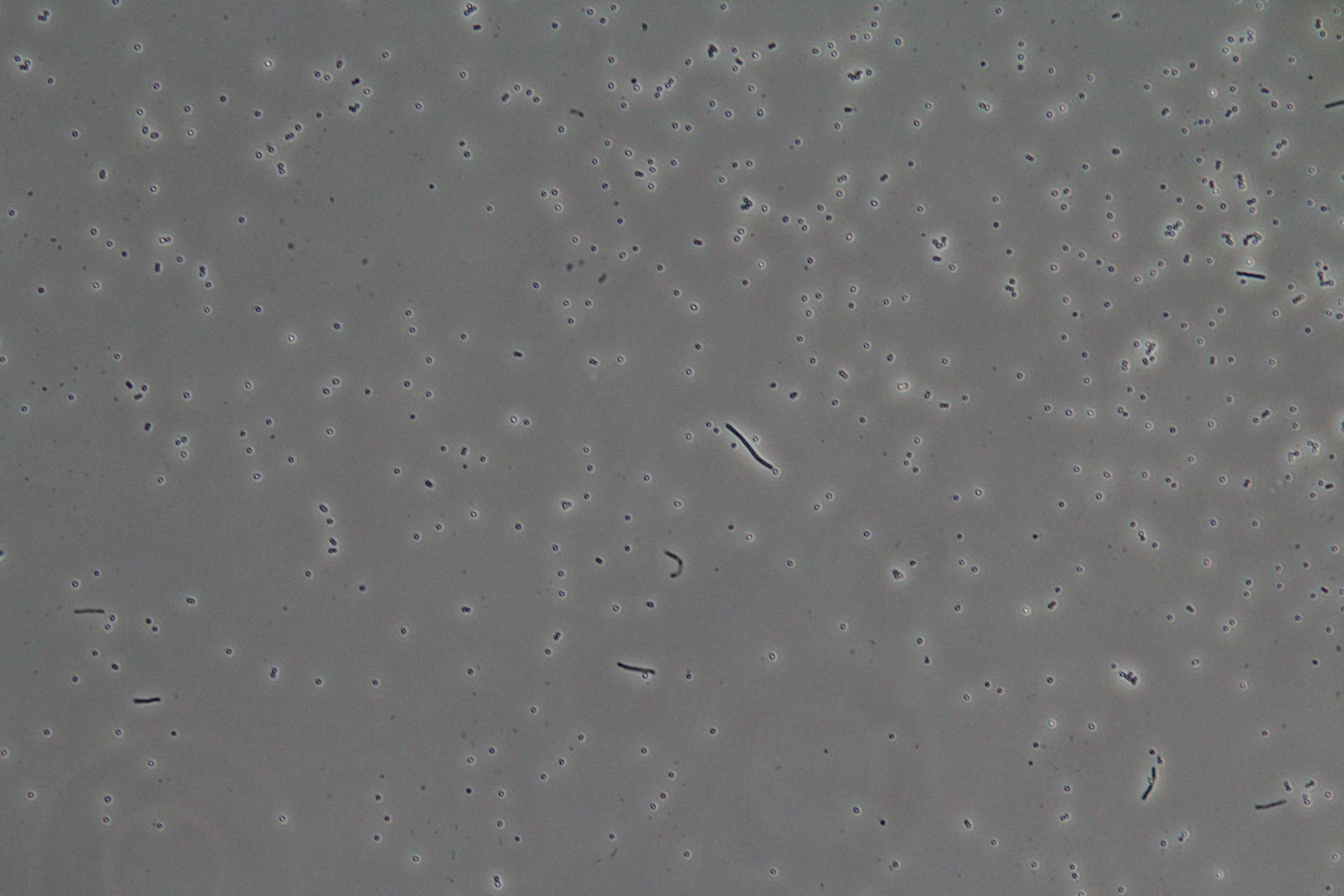
| Figure 1. Cluster of WT vegetative Bacillus (Phase Contrast) | Figure 2. Spores (circular) and vegetative (rod-shaped) Bacillus (Phase Contrast) |
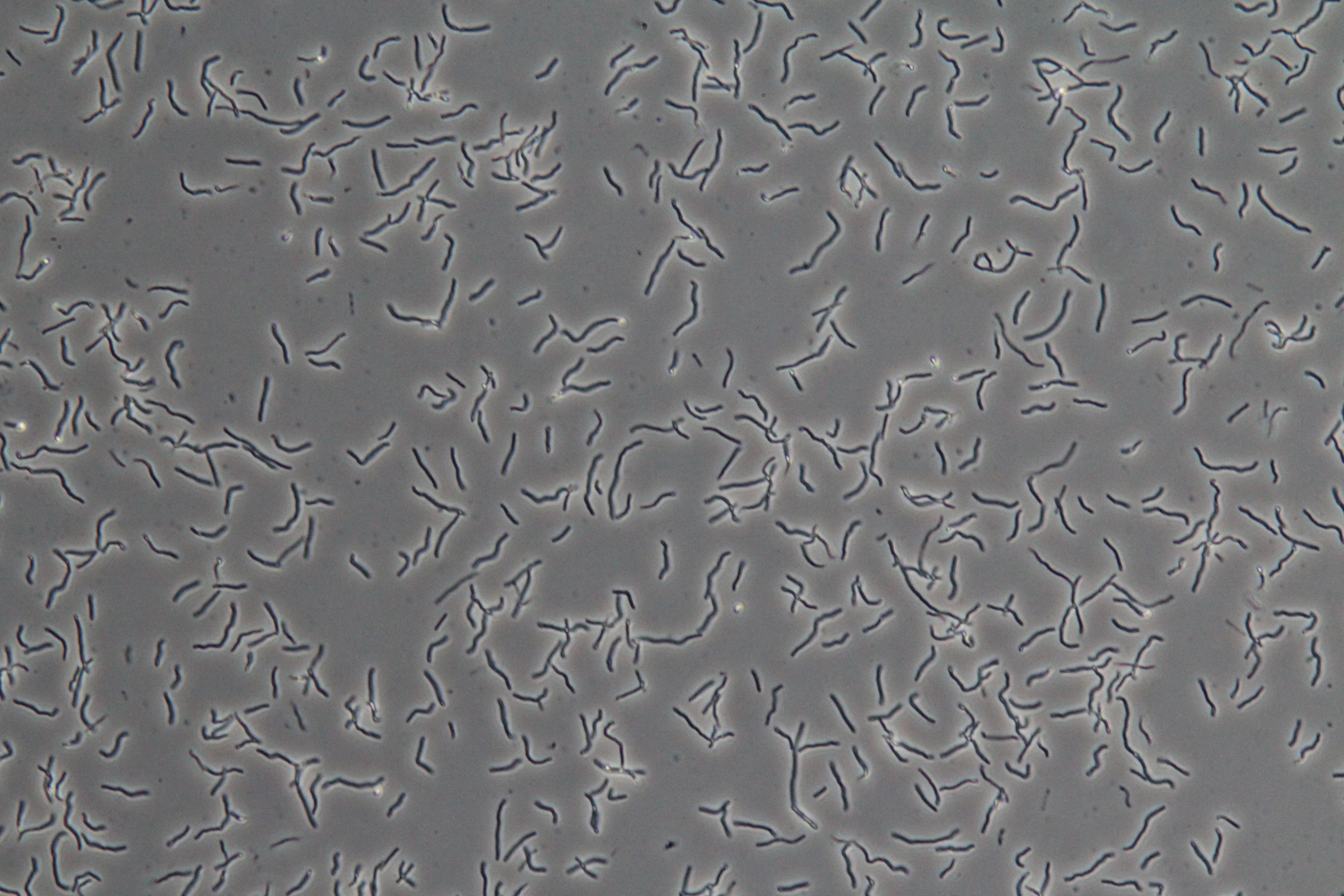
| 
|
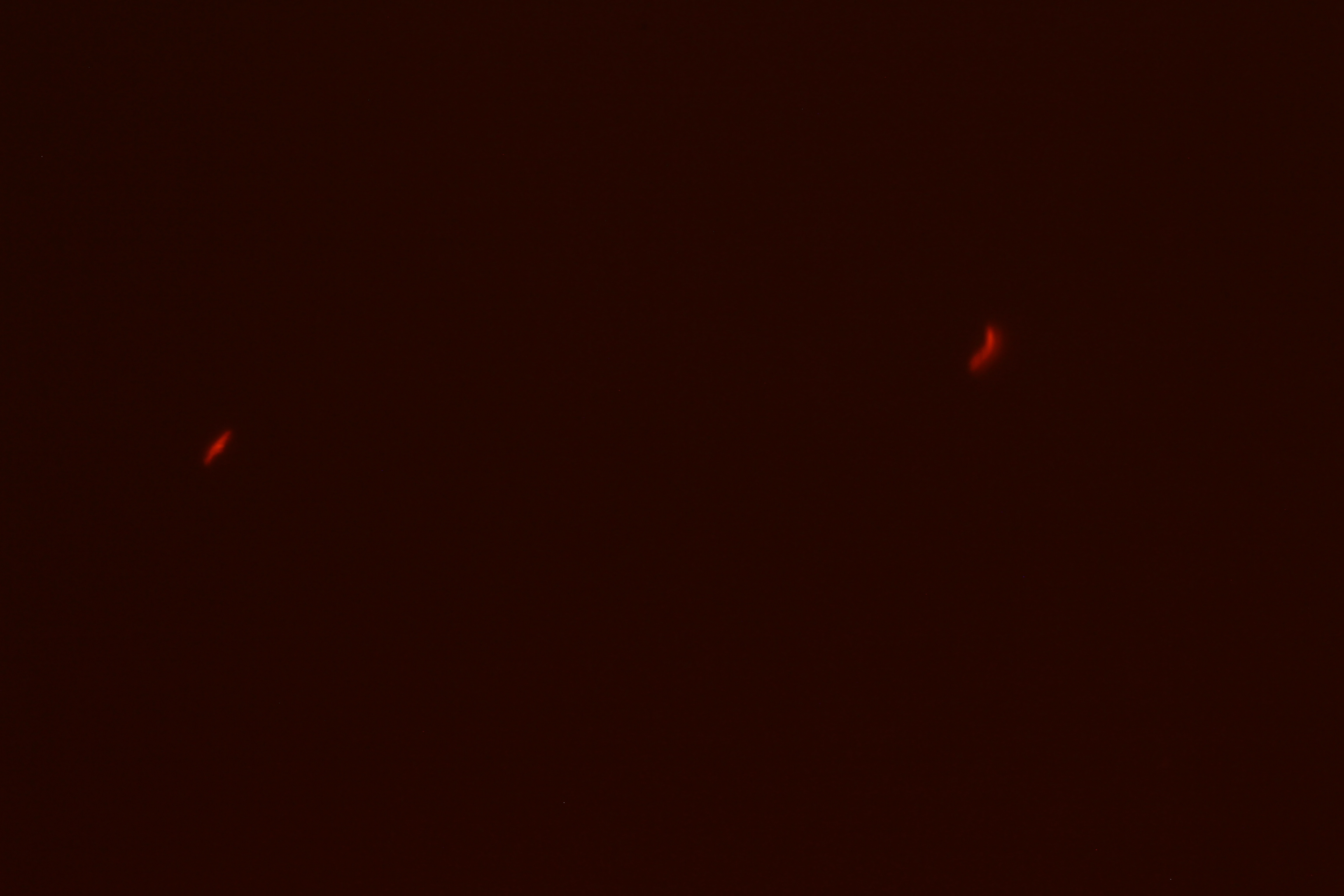
| Figure 3. SacY + RFP Bacillus with RFP linked to sucrose inducer promoter (Phase Contrast) | Figure 4. SacY + RFP Bacillus with RFP linked to sucrose inducer promoter (Fluorescence) |
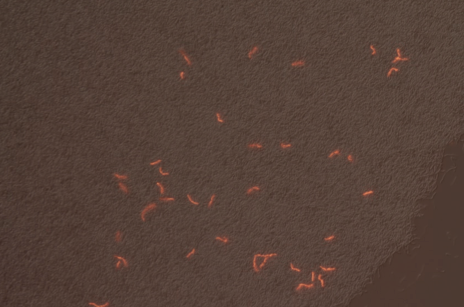
|
| Figure 7. Overlay of Figure 5 and 6 |
Conclusions
Through this project, we constructed and synthesized prototype chromogenic biosensors to detect sucrose metabolism and sporulation in Bacillus subtilis. Fluorescent micrographs show that our transformations were successful and that our engineered biosensors fluoresce. However, we still have further work to do in order to isolate the fluorescent colonies and characterize our promoters of interest, which may be exhibiting promoter leakiness. This future work will be important to prepare for the 2016 EuCROPIS satellite mission and a helium balloon test next year.
BioBricks
[http://parts.igem.org/Part:BBa_K1218001 BBa_K1218001]: This part is the sacY promoter; sacY is a gene that regulates sucrose induction in Bacillus subtilis.
[http://parts.igem.org/Part:BBa_K1218020 BBa_K1218020]: This is a compound part containing the sacY promoter (BBa_K1218001: a sucrose inducer in Bacillus subtilis) and a ribosome binding site (BBa_B0034).
[http://parts.igem.org/Part:BBa_K1218021 BBa_K1218021]: This part is the spo0A promoter; spo0A regulates sporulation in Bacillus subtilis.
[http://parts.igem.org/Part:BBa_K1218023 BBa_K1218023]: This composite part contains sacY (BBa_K1218001: a sucrose inducer in Bacillus subtilis), a ribosome binding site (BBa_B0034), and eForRed, a red chromoprotein developed by Uppsala (BBa_K592012). We intend to use it as a chromogenic reporter induced by sucrose.
[http://parts.igem.org/Part:BBa_K1218025 BBa_K1218025]: This composite part contains sacY (BBa_K1218001: a sucrose inducer in Bacillus subtilis), a ribosome binding site (BBa_B0034), and a red fluorescent protein (BBa_E1010). We intend to use it as a chromogenic reporter induced by sucrose.
Acknowledgements
We would like to thank:
- Ryan Kent, Dr. Lynn Rothschild, Dr. Joe Shih, and Dr. Kosuke Fujisima -- Primary advisors
- Dr. Gary Wessel — Advice on testing construct
- Dr. Rocco Mancinelli — Feedback on our presentation, papers about the EuCROPIS mission
- Dr. Lilah Rahn-Lee — Advice on choosing B. subtilis genes
- Dr. Daniel R. Zeigler, BGSC Director and the Bacillus Genetic Stock Center — Free strains of Bacillus integration vectors
- Dr. Elwood Agasid — Explanation of EuCROPIS mission from an engineering perspective
References
- Chibazakura, T., Kawamura, F., Takahashi, H. (1991). Differential Regulation of spo0A Transcription in Bacillus subtilis: Glucose Represses Promoter Switching at the Initiation of Sporulation. Journal of Bacteriology 173: 2625-2632.
- Crutz, A., Steinmetz, M. (1992). Transcription of the Bacillus subtilis sacX and sacY Genes, Encoding Regulators of Sucrose Metabolism, Is Both Inducible by Sucrose and Controlled by the DegS-DegU Signalling System. Journal of Bacteriology 174: 6087-6095.
- Hauslage, J., Lebert, M., Strauch, S. M., Mancinelli, R. Eu:CROPIS - Euglena: Combined Regenerative Organic-Food Production In Space. Space Definition Document.
- Horneck, G. (1992). Responses of Bacillus Subtilis spores to space environment: Results from experiments in space. Microbiology and Molecular Biology Reviews. Origins of Life and Evolution of the Biosphere 23: 37-52.
- Horneck, G., Klaus, D. M., Mancinelli, R. L. (2010). Space Microbiology. Microbiology and Molecular Biology Reviews 74: 121-156.
- Keijser, B. J. F., Beek, A. T., Rauwerda, H., Schuren, F., Montijn, R., Van der Spek, H., Brul, S. (2007). Analysis of Temporal Gene Expression during "Bacillus subtilis" Spore Germination and Outgrowth. Journal of Bacteriology 189: 3624-3634.
 "
"