Team:Clemson/Project
From 2013.igem.org
(→Project Details) |
|||
| (10 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
FDA has maintained a zero-tolerance policy for several foodborne pathogens. For example, a policy of “zero-tolerance” for ''Listeria monocytogenes'' in ready-to-eat foods means that the detection of any ''L. monocytogenes'' in either of two 25 gram samples of a food renders the food adulterated; the infectious dosage of ''E. coli'' O157:H7 has been determined to be 10 cells; the Environmental Protection Agency standard for ''E. coli'' O157:H7 in water is 40 cells per liter. The current detection methods suffer from one or more of the following limitations: 1) the requirement of pre-enrichment and enrichment to increase the number of target pathogens, e.g., bio-chemical assays and immunoassays, 2) high detection limit, e.g., 10^3 – 10^5 CFU per ml or per gram of sample for immunoassays, 3) inability to distinguish viable from non-viable cells, e.g., PCR-based detection methods, 4) small sample volume capacity, e.g., microfluidic-based biosensors (µl instead of the required ml to liter capacity), 5) tedious detection procedures, and 6) the current high per-assay cost. | FDA has maintained a zero-tolerance policy for several foodborne pathogens. For example, a policy of “zero-tolerance” for ''Listeria monocytogenes'' in ready-to-eat foods means that the detection of any ''L. monocytogenes'' in either of two 25 gram samples of a food renders the food adulterated; the infectious dosage of ''E. coli'' O157:H7 has been determined to be 10 cells; the Environmental Protection Agency standard for ''E. coli'' O157:H7 in water is 40 cells per liter. The current detection methods suffer from one or more of the following limitations: 1) the requirement of pre-enrichment and enrichment to increase the number of target pathogens, e.g., bio-chemical assays and immunoassays, 2) high detection limit, e.g., 10^3 – 10^5 CFU per ml or per gram of sample for immunoassays, 3) inability to distinguish viable from non-viable cells, e.g., PCR-based detection methods, 4) small sample volume capacity, e.g., microfluidic-based biosensors (µl instead of the required ml to liter capacity), 5) tedious detection procedures, and 6) the current high per-assay cost. | ||
The aim of this project is develop a Universal Self-Amplified ('''USA''') Biosensor that addresses the aforementioned disadvantages of current detection methods. This two component system utilizes a universal signal amplification bacterial system and a unique pathogen-specific detection counterpart for a one-step detection of target microorganisms in a scalable volume. | The aim of this project is develop a Universal Self-Amplified ('''USA''') Biosensor that addresses the aforementioned disadvantages of current detection methods. This two component system utilizes a universal signal amplification bacterial system and a unique pathogen-specific detection counterpart for a one-step detection of target microorganisms in a scalable volume. | ||
| - | |||
| - | |||
{| | {| | ||
| - | [[File: | + | [[File:wikisystem.png|800px|frameless|center]] |
|} | |} | ||
Our universal self-amplified (USA) biosensor is composed of two parts. The first part is a phage adaptor specific for each pathogen that can be identified. The phage would deliver an AHL signal sending DNA construct into any pathogen present. The second part is our USA biosensor. The USA biosensor receives the AHL signal, self-amplifies it, and puts out an easily detectable signal such as GFP. This is universal in the sense that it would amplify the AHL signal from any pathogen-specific phage adaptor. We have not yet constructed a phage delivery system, so our current system utilizes a “model pathogen” harboring a plasmid with the AHL signal sender. This model pathogen represents a bacterium that has been infected by the phage adaptor. | Our universal self-amplified (USA) biosensor is composed of two parts. The first part is a phage adaptor specific for each pathogen that can be identified. The phage would deliver an AHL signal sending DNA construct into any pathogen present. The second part is our USA biosensor. The USA biosensor receives the AHL signal, self-amplifies it, and puts out an easily detectable signal such as GFP. This is universal in the sense that it would amplify the AHL signal from any pathogen-specific phage adaptor. We have not yet constructed a phage delivery system, so our current system utilizes a “model pathogen” harboring a plasmid with the AHL signal sender. This model pathogen represents a bacterium that has been infected by the phage adaptor. | ||
| Line 14: | Line 12: | ||
== Results == | == Results == | ||
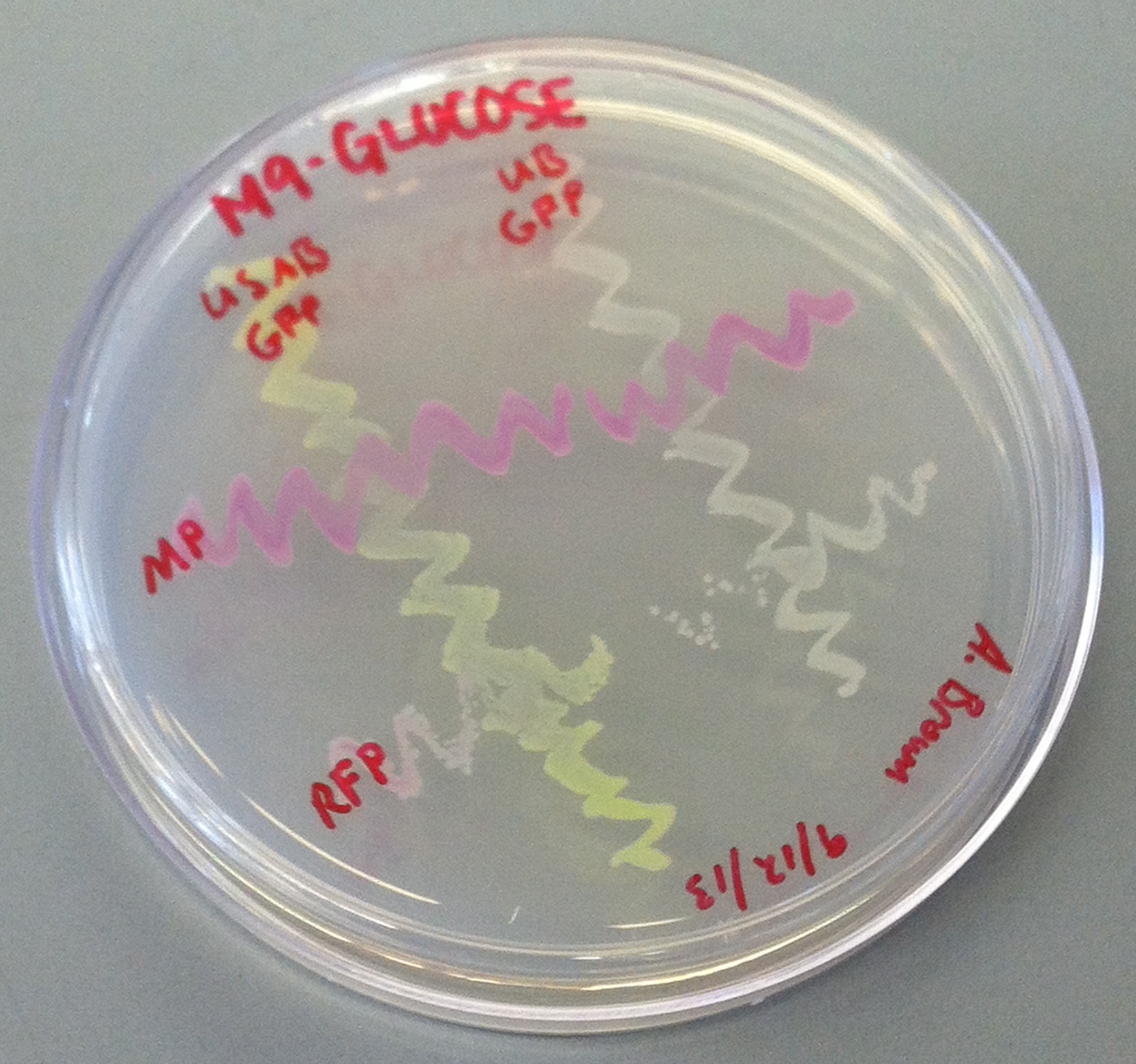
[[File:CUPlate.png|400px|thumb|center|Streak plate showing all four constructs]] | [[File:CUPlate.png|400px|thumb|center|Streak plate showing all four constructs]] | ||
| + | {| | ||
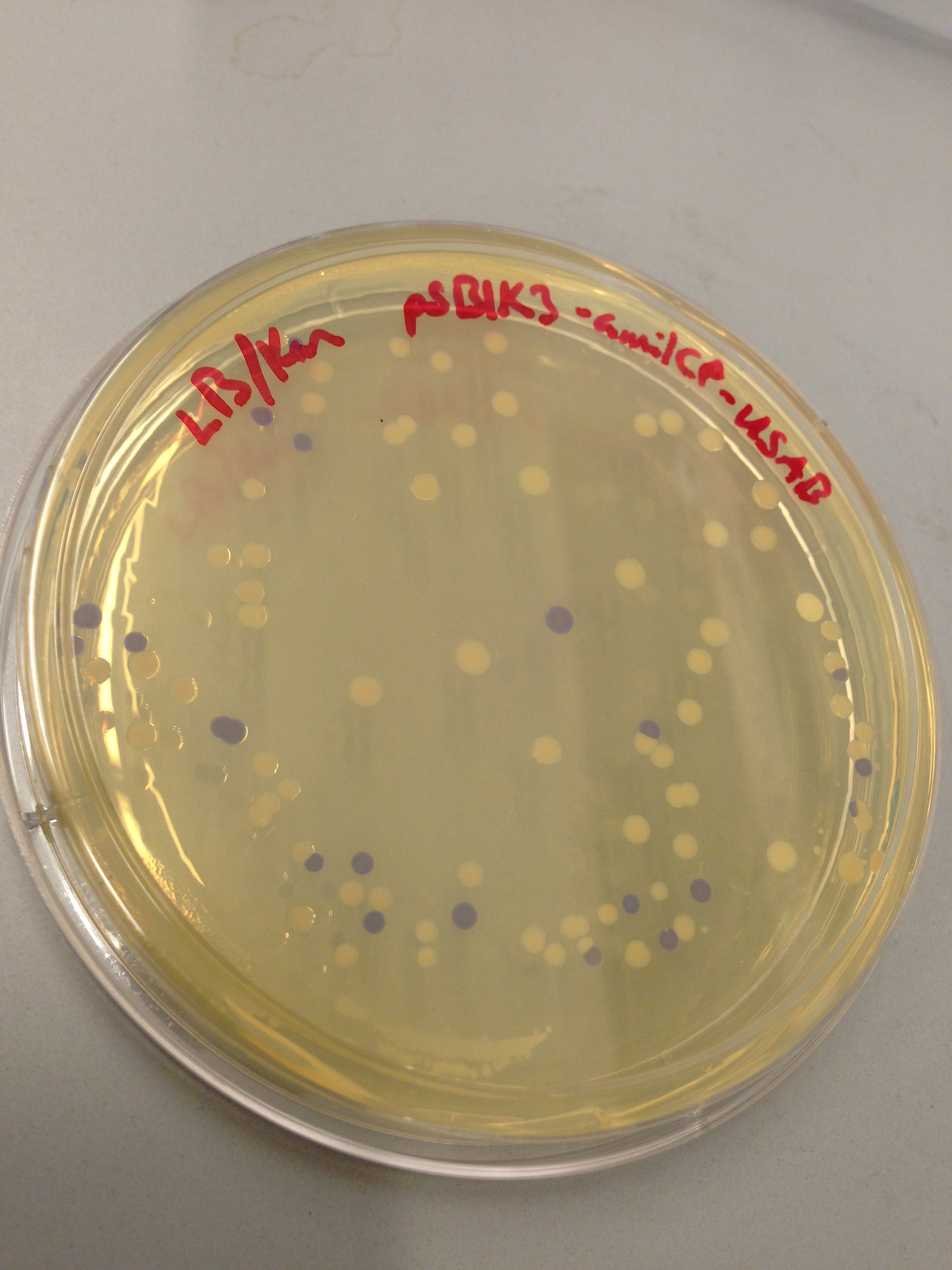
| + | [[File:blueplate.png|400px|thumb|center|Transformation plate showing amilCP expression]] | ||
{| | {| | ||
''E. coli'' DH10B cultures hosted all the plasmid constructs. A total of four cultures were used: the universal sef-amplified biosensor (USAB), the universal non-self-amplified biosensor (UB), the “model pathogen” (MP), and the negative control non-AHL-producing “model pathogen” (RFP). See above for details on the plasmid constructs of each culture. | ''E. coli'' DH10B cultures hosted all the plasmid constructs. A total of four cultures were used: the universal sef-amplified biosensor (USAB), the universal non-self-amplified biosensor (UB), the “model pathogen” (MP), and the negative control non-AHL-producing “model pathogen” (RFP). See above for details on the plasmid constructs of each culture. | ||
Latest revision as of 00:49, 29 October 2013
Overall project
FDA has maintained a zero-tolerance policy for several foodborne pathogens. For example, a policy of “zero-tolerance” for Listeria monocytogenes in ready-to-eat foods means that the detection of any L. monocytogenes in either of two 25 gram samples of a food renders the food adulterated; the infectious dosage of E. coli O157:H7 has been determined to be 10 cells; the Environmental Protection Agency standard for E. coli O157:H7 in water is 40 cells per liter. The current detection methods suffer from one or more of the following limitations: 1) the requirement of pre-enrichment and enrichment to increase the number of target pathogens, e.g., bio-chemical assays and immunoassays, 2) high detection limit, e.g., 10^3 – 10^5 CFU per ml or per gram of sample for immunoassays, 3) inability to distinguish viable from non-viable cells, e.g., PCR-based detection methods, 4) small sample volume capacity, e.g., microfluidic-based biosensors (µl instead of the required ml to liter capacity), 5) tedious detection procedures, and 6) the current high per-assay cost. The aim of this project is develop a Universal Self-Amplified (USA) Biosensor that addresses the aforementioned disadvantages of current detection methods. This two component system utilizes a universal signal amplification bacterial system and a unique pathogen-specific detection counterpart for a one-step detection of target microorganisms in a scalable volume.
Our universal self-amplified (USA) biosensor is composed of two parts. The first part is a phage adaptor specific for each pathogen that can be identified. The phage would deliver an AHL signal sending DNA construct into any pathogen present. The second part is our USA biosensor. The USA biosensor receives the AHL signal, self-amplifies it, and puts out an easily detectable signal such as GFP. This is universal in the sense that it would amplify the AHL signal from any pathogen-specific phage adaptor. We have not yet constructed a phage delivery system, so our current system utilizes a “model pathogen” harboring a plasmid with the AHL signal sender. This model pathogen represents a bacterium that has been infected by the phage adaptor.
Results
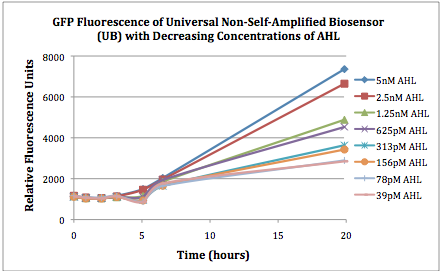
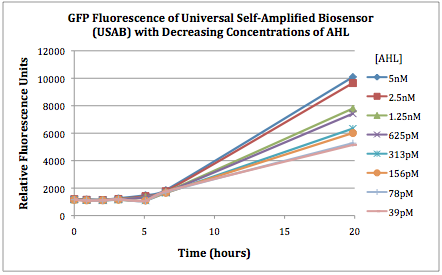
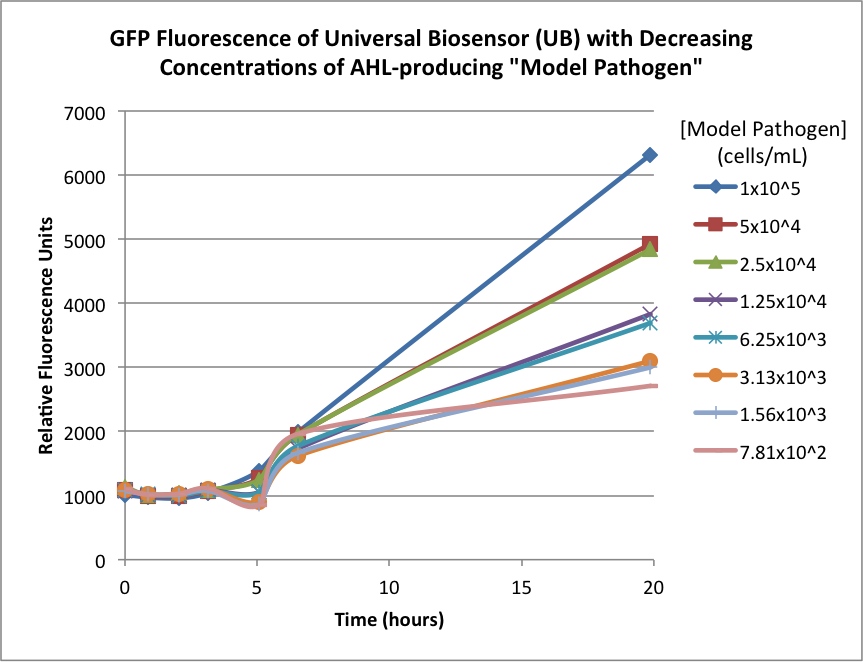
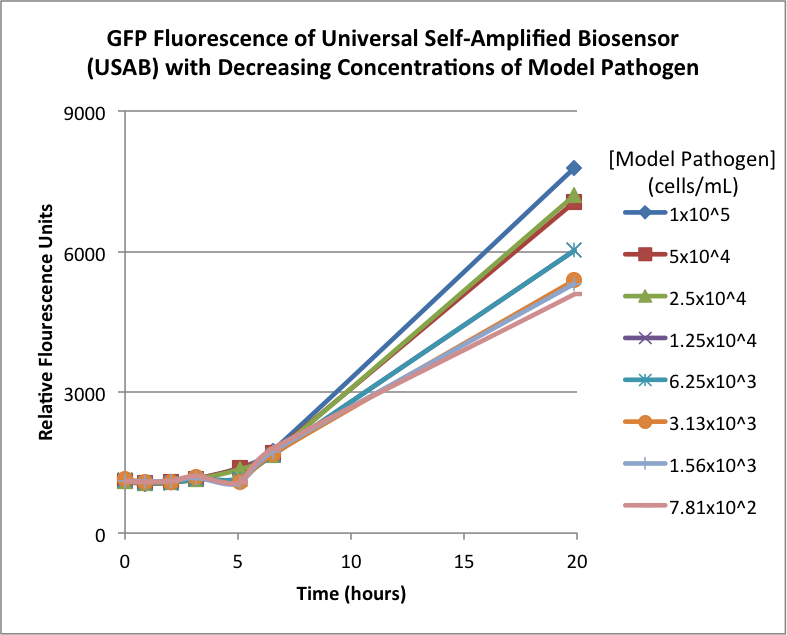
Figures 2A and 2B show the response of the UB and USAB, respectively, to different concentrations of the “model pathogen.” As expected, higher concentrations of the “model pathogen,” which produces AHL, cause relatively higher GFP emission over time.
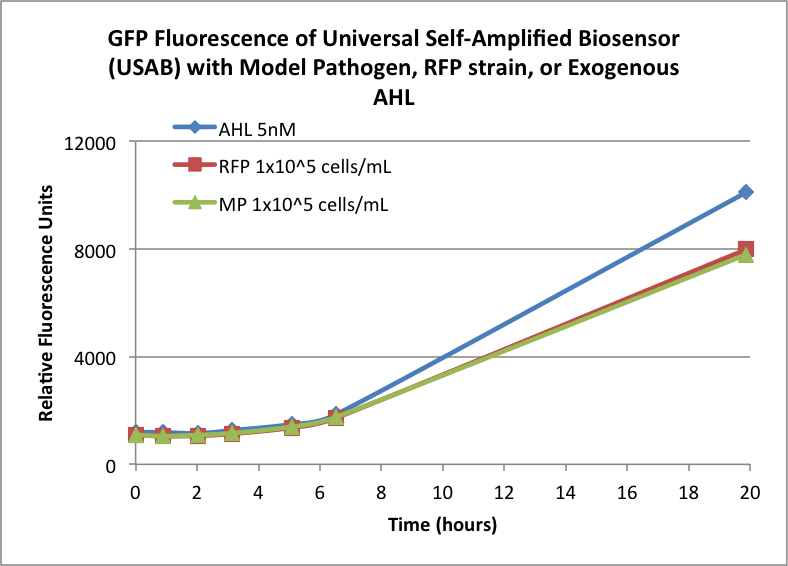
Figures 3A and 3B show the response of the UB and USAB, respectively, to AHL (5nM), “model pathogen” (1x10^5 cells/mL), or RFP strain (the non-AHL-producing model pathogen at 1x10^5 cells/mL). Unexpectedly, the relative GFP emission appears to increase over time regardless of whether the strain present produces AHL. This could mean the fluorescence is increasing only as the biosensor strain grows, with GFP expression per cell remaining relatively constant. However, the presence of AHL does seem to increase the slope of fluorescence.
 "
"