Team:DTU-Denmark/Notebook/10 July 2013
From 2013.igem.org
(→Results) |
|||
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:DTU-Denmark/Templates/StartPage|}} | + | {{:Team:DTU-Denmark/Templates/StartPage|10 July 2013}} |
| - | + | Navigate to the [[Team:DTU-Denmark/Notebook/9_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/11_July_2013|Next]] Entry | |
| - | + | =Lab 208= | |
| - | =208= | + | <hr/> |
==Main purpose== | ==Main purpose== | ||
| + | <hr/> | ||
Run gel with 9 PCR samples of Nir operon from ''Pseudomonas aeruginosa'' | Run gel with 9 PCR samples of Nir operon from ''Pseudomonas aeruginosa'' | ||
| Line 11: | Line 12: | ||
==Who was in the lab== | ==Who was in the lab== | ||
| - | Ariadni,Henrike,Julia,Natalia | + | <hr/> |
| + | Ariadni, Henrike, Julia, Natalia | ||
==Procedure== | ==Procedure== | ||
| - | + | <hr/> | |
===Run gel=== | ===Run gel=== | ||
* 3 samples from PCR on the 09.07.2013 using PAO1 as template source | * 3 samples from PCR on the 09.07.2013 using PAO1 as template source | ||
| Line 95: | Line 97: | ||
==Results== | ==Results== | ||
| - | + | <hr/> | |
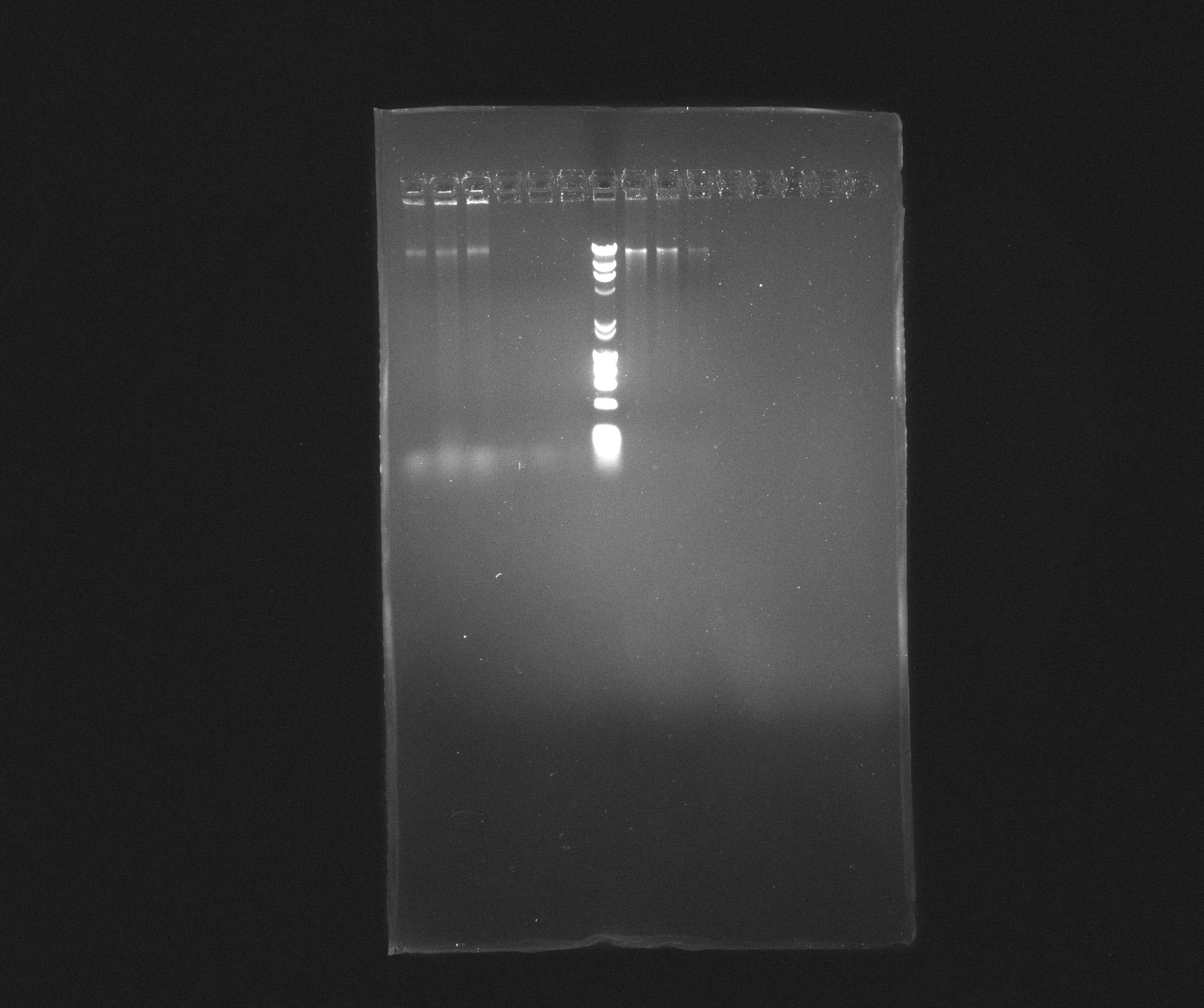
0.8 % Agorase Gel (Nir operon) | 0.8 % Agorase Gel (Nir operon) | ||
| Line 102: | Line 104: | ||
*2: Nir, yesterday's PCR, source: culture PAO1 | *2: Nir, yesterday's PCR, source: culture PAO1 | ||
*3: Nir, yesterday's PCR, source: culture PAO1 | *3: Nir, yesterday's PCR, source: culture PAO1 | ||
| - | *4: Nir, yesterday's PCR, source: purification from | + | *4: Nir, yesterday's PCR, source: purification from 09.07.2013 |
| - | *5: Nir, yesterday's PCR, source: purification from | + | *5: Nir, yesterday's PCR, source: purification from 09.07.2013 |
| - | *6: Nir, yesterday's PCR, source: purification from | + | *6: Nir, yesterday's PCR, source: purification from 09.07.2013 |
*7: Broadband Ladder | *7: Broadband Ladder | ||
| - | *8: Nir | + | *8: Nir purification of 09.07.2013 |
| - | *9: Nir | + | *9: Nir purification of 09.07.2013 |
| - | *10: Nir | + | *10: Nir purification of 09.07.2013 |
[[File:2013_07_10_gel_nir.jpg |600px]] | [[File:2013_07_10_gel_nir.jpg |600px]] | ||
==Comments== | ==Comments== | ||
| + | <hr/> | ||
Kanamycin plates were of bad quality | Kanamycin plates were of bad quality | ||
| Line 120: | Line 123: | ||
==Conclusions== | ==Conclusions== | ||
| + | <hr/> | ||
Nir forms a nice, but weak, band in most samples but the fragments are still far longer than expected. It looks like they are somewhere around 16 kb where we rather expected 9102 bp. We will try to cut down the extension time and see if we can get something out of it. | Nir forms a nice, but weak, band in most samples but the fragments are still far longer than expected. It looks like they are somewhere around 16 kb where we rather expected 9102 bp. We will try to cut down the extension time and see if we can get something out of it. | ||
Latest revision as of 20:40, 16 September 2013
10 July 2013
Contents |
Lab 208
Main purpose
Run gel with 9 PCR samples of Nir operon from Pseudomonas aeruginosa
Transformation of Biobricks
PCR reaction for Nir operon
Who was in the lab
Ariadni, Henrike, Julia, Natalia
Procedure
Run gel
- 3 samples from PCR on the 09.07.2013 using PAO1 as template source
- 3 samples from PCR on the 09.07.2013 using PCR purification of 08.07.2013 as template source
- ladder broad range biggest fragment = 20 kb
- 3 samples from purification of 09.07.2013
Transformation of Biobricks
According to the iGEM protocol ([http://parts.igem.org/Help:Protocols/Transformation transformation protocol]) with slight changes.
- step 1: 100 μl cells
- step 2: 1.5 μl of the resuspended DNA
- step 6: 90 sec (instead of 60 s)at 42 oC
- step 9: Incubation without shaking
Transformation list
| BioBrick number | Backbone | Resistance | Plate | Well | Copies |
|---|---|---|---|---|---|
| J04450 | pSB3C5 | CAM | 2 | 4D | 10-12 |
| J04450 | pSB4C5 | CAM | 2 | 4F | 5 |
| J04450 | pSB4K5 | Kan | 2 | 6H | 5 |
| J04450 | pSB1T3 | Tetra | 2 | 8B | 100-300 |
| J04450 | pSB3T5 | Tetra | 2 | 8D | 10-12 |
| J04450 | pSB1AK3 | Amp+Kan | 2 | 12B | 100-300 |
| J04450 | pSB1A3 | Amp | 2 | 2H | 100-300 |
| J04450 | pSB1A2 | Amp | 5 | 1B | 100-300 |
| J04450 | pSB6A1 | Amp | 5 | 1K | 10-15 |
| J04450 | pSB1K3 | Kan | 5 | 5A | 100-300 |
| J04450 | pSB3K3 | Kan | 5 | 5E | 10-12 |
| K325909 | pSB1C3 | CAM | 1 | 4L | 100-300 |
| K325209 | pSB1C3 | CAM | 1 | 12H | 100-300 |
| K325219 | pSB1C3 | CAM | 1 | 2D | 100-300 |
| J04421 | pSB1C3 | CAM | 3 | 15O | 100-300 |
| E0430 | pSB1C3 | CAM | 3 | 20K | 10-300 |
Extraction PCR
9 samples from culture pAO1 10 reactions
- dNTP: 1 ul (10 ul)
- HF buffer: 10 ul (100 ul)
- Phusion polymerase: 0.5 ul (5 ul)
- H2O: 31.5 ul (315 ul)
Settings for PCR with three different elongation times
| Temperature (oC) | Time (min) | Rounds |
|---|---|---|
| 98 | 2:00 | 1 |
| 98 | 0:20 | 35 |
| 66 | 0:45 | 35 |
| 72 | 2:00/3:00/4:00 | 35 |
| 72 | 5:00 | 1 |
| 4 | ∞ | - |
Results
0.8 % Agorase Gel (Nir operon)
Wells
- 1: Nir, yesterday's PCR, source: culture PAO1
- 2: Nir, yesterday's PCR, source: culture PAO1
- 3: Nir, yesterday's PCR, source: culture PAO1
- 4: Nir, yesterday's PCR, source: purification from 09.07.2013
- 5: Nir, yesterday's PCR, source: purification from 09.07.2013
- 6: Nir, yesterday's PCR, source: purification from 09.07.2013
- 7: Broadband Ladder
- 8: Nir purification of 09.07.2013
- 9: Nir purification of 09.07.2013
- 10: Nir purification of 09.07.2013
Comments
Kanamycin plates were of bad quality
Tetracyclin plates were made by adding 22.5 uL of tetracyclin stock to LB plates
- Stock tetracyclin: 10 mg/mL
- Final concentration in plates: 15 ug/mL
Conclusions
Nir forms a nice, but weak, band in most samples but the fragments are still far longer than expected. It looks like they are somewhere around 16 kb where we rather expected 9102 bp. We will try to cut down the extension time and see if we can get something out of it.
Navigate to the Previous or the Next Entry
 "
"