Team:Heidelberg/Templates/Modelling/Ind-Production
From 2013.igem.org
m |
m |
||
| (20 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<div class="container"><div class="row" style="float:left"><div class="col-sm-12"> | <div class="container"><div class="row" style="float:left"><div class="col-sm-12"> | ||
<h2 id="Challenge"> Challenge </h2> | <h2 id="Challenge"> Challenge </h2> | ||
| - | A challenge we had to face during the <a href="https://2013.igem.org/Team:Heidelberg/Project/Tag-Optimization" | + | A challenge we had to face during the <a href="https://2013.igem.org/Team:Heidelberg/Project/Tag-Optimization">characterization and optimization of indC</a> was to identify the production kinetics of indigoidine. |
| - | In order to disentangle the underlying mechanisms of bacterial growth and peptide synthesis, we decided to set up a mathematical model based on coupled ordinary differential equations (ODEs). Calibrated with our experimental time-resolved data, the mathematical model could potentially not only elucidate how | + | In order to disentangle the underlying mechanisms of bacterial growth and peptide synthesis, we decided to set up a mathematical model based on coupled ordinary differential equations (ODEs). Calibrated with <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/3/38/Heidelberg_IndPD_Fig11.png" title="<b>Figure 8: Indigoidine Production Varies Among Engineered Indigoidine Synthetases.</b> |
| + | The absorption spectrum of liquid cultures expressing engineered variants of the indC indigoidine synthetase were measured for 30 hours. The graphs show the OD590 of indigoidine in <i>E. coli</i> liquid cultures over time. The absorption of the cell suspension has been subtracted (see OD Measurement in the Methods section). The red graph relates to an <i>E. coli</i> TOP10 negative control, the green graph depicts the indigoidine production of an indC variant, in which the T-domain has been exchanged with our synthetic T-domain #3, whereas the blue graphs refers to synthetic T-domain #4. | ||
| + | The amount of indigoidine reaches a maximum before it drops again. This is due the instability of indigoidine, which is reduced to its fluorescent leuco-form under the influence of reducing agents, light and high temperature. The local absorption maximum at 590 nm (keto-indigoidine) decreases after 15-25 hours, whereas the absorption maximum at 430 nm (leuco-indigoidine) increases (data not shown). Note that the maximum level of indigoidine differs among the engineered indigoidine synthetases.">our experimental time-resolved data</a>, the mathematical model could potentially not only elucidate how indigoidine production influences growth of bacteria but also provide a more quantitative understanding of the synthesis efficiency of <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/4/4d/Heidelberg_IndPD_Fig10.png" caption="<b>Figure 8: Indigoidine Production Depend on Combination of T-domain and PPTase</b> | ||
| + | The absorption spectrum of liquid cultures expressing engineered variants of the indC indigoidine synthetase were measured for 30 hours. The graphs show the OD590 of indigoidine in <i>E. coli</i> liquid cultures over time. The absorption of the cell suspension has been subtracted (see OD Measurement in the Methods section). The left diagram shows the indigoidine production over time of a liquid culture expressing an engineered indigoidine synthetase with the synthetic T-domain #1 (synT1) and a PPTase, whereas each graph refers to a specific PPTase coexpressed. The diagram on the right shows the corresponding data for the indigoidine synthetase with the synthetic T-domain #3 (synT3). | ||
| + | The indigoidine production correlates to the combination of a T-domain with a respective PPTase and the PPTases vary in their ability to activate T-domains. For example, DelC is unable to activate synT 1 but activates synT3, whereas svp activates synT1 but is unable to activate synT3. Note also, that activation of synT3 by EntD results in the highest amount of indigoidine production among all the combinations shown.">the different T domains and PPTases</a> that were tested. | ||
<h2 id="Approach"> Approach </h2> | <h2 id="Approach"> Approach </h2> | ||
<p> | <p> | ||
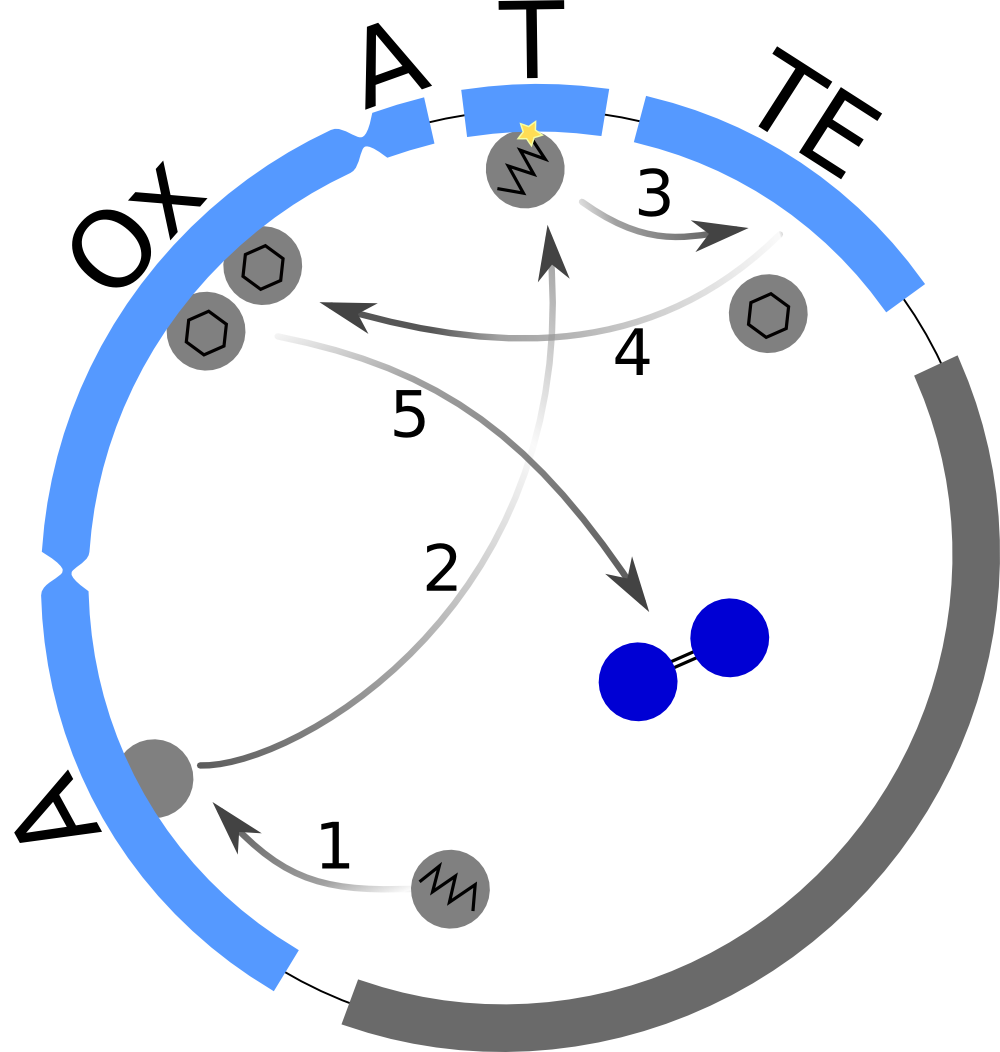
| - | First, we set up a mind model based on the fact that the functional | + | First, we set up a mind model based on the fact that the functional indigoidine dimer is produced from two glutamines (Glu) that are each cyclized (cGlu)[1] (<a class="fancybox fancyFigure" href="/wiki/images/8/83/Heidelberg_IndSynthesis_scheme.png" title="<b>Figure 1: The indigoidine synthetase indC produces a blue pigment and is used for the indigoidine-tag</b>. After activation of L-glutamine by the A domain and its binding to the T domain, it is cyclized by the TE domain and oxidized by the Ox domain. Dimerization is thought to happen by air.[1]">Fig. 1</a>), and our observation that indigoidine (Ind)-producing bacteria (Bac) <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/c/cf/Heidelberg_IndPD_Fig6.png" caption="<b>Figure 3: Comparison between different <em>E. coli</em> strains and PPTases:</b> |
| + | <b>a)</b> Comparison of different <em>E. coli</em> strains examining growth and indigoidine production | ||
| + | The figure shows five different strains of <em>E. coli</em> that have been co-transformed with an indC expression plasmid and a sfp expression plasmid. The negative control is <em>E. coli</em> TOP10 without a plasmid. All transformants have been grown on LB agar for 48 hours at room temperature, cells were not induced. One can see that even without induction all strains express the indigoidine synthetase and produce the blue pigment indigoidine. However, the strains BAP1 and NEB Turbo grow faster in the first day, exhibiting a white phenotype (data not shown). Colonies on the plate of <em>E. coli</em> TOP10 are very small and dark blue/ black. Assuming that indigoidine production inhibits cell growth due to its toxicity, we concluded that TOP10 produced the most indigoidine among the strains we tested. We used <em>E. coli</em> TOP10 for the following experiments. | ||
| + | <b>b)</b> Comparison between different PPTases concerning overall indigoidine production | ||
| + | The Figure shows <i>E. coli</i> TOP10 cells co-transformed with indC and four different PPTases (sfp, svp, entD and delC), respectively. The image bottom left shows <i>E. coli</i> TOP10 cells without additional PPTase and the negative control is TOP10 without a plasmid. | ||
| + | ">grow slower</a> than mock controls. Those hypotheses resulted in a general model scheme depicting the interdependency between indigoidine synthesis and bacterial growth. With the mathematical model, we could then validate whether there is indeed a negative feedback from the indigoidine production to the growth of bacteria. | ||
</p> | </p> | ||
<center> | <center> | ||
| - | <a class="fancybox fancyFigure" href="/wiki/images/8/83/Heidelberg_IndSynthesis_scheme.png" title="<b>Figure 1: The indigoidine synthetase indC produces a blue pigment and is used for the indigoidine-tag</b>. After activation of L-glutamine by the A domain and its binding to the T domain, it is cyclized by the TE domain and oxidized by the Ox domain. Dimerization is thought to happen by air."> | + | <a class="fancybox fancyFigure" href="/wiki/images/8/83/Heidelberg_IndSynthesis_scheme.png" title="<b>Figure 1: The indigoidine synthetase indC produces a blue pigment and is used for the indigoidine-tag</b>. After activation of L-glutamine by the A domain and its binding to the T domain, it is cyclized by the TE domain and oxidized by the Ox domain. Dimerization is thought to happen by air.[1]"> |
<img style="width:30%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/8/83/Heidelberg_IndSynthesis_scheme.png"/> | <img style="width:30%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/8/83/Heidelberg_IndSynthesis_scheme.png"/> | ||
| - | <figcaption style="width:60%;"><b>Figure 1: The indigoidine synthetase indC produces a blue pigment and is used for the indigoidine-tag</b>. After activation of L-glutamine by the A domain and its binding to the T domain, it is cyclized by the TE domain and oxidized by the Ox domain. Dimerization is thought to happen by air.</figcaption> | + | <figcaption style="width:60%;"><b>Figure 1: The indigoidine synthetase indC produces a blue pigment and is used for the indigoidine-tag</b>. After activation of L-glutamine by the A domain and its binding to the T domain, it is cyclized by the TE domain and oxidized by the Ox domain. Dimerization is thought to happen by air.[1]</figcaption> |
</a> | </a> | ||
</center> | </center> | ||
<p> | <p> | ||
| - | Since we had already established our quantitative | + | Since we had already established our quantitative indigoidine production assay (see <a href="https://2013.igem.org/Team:Heidelberg/Project/Tag-Optimization">Tag-Optimization</a>) in a time-dependent manner, we wanted to further exploit these experimental data via quantitative dynamic modeling. The change of bacteria and Indigoidine with time was measured via <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/6/64/Heidelberg_IndPD_Fig5.png" caption="<b>Figure 13: Quantification of dye in cellular culture by OD measurements at robust and sensitive wavelengths. </b>The contribution of the scattering by the cellular components at the sensitive wavelength, i.e. 590 nm for indigoidine has to be subtracted from the overall OD at this wavelength. For a detailed description of the calculation refer to text below. |
| - | Such ODEs contain parameters that characterize e.g. growth or synthesis rates for bacteria or | + | Figure adopted from [15]">optical density</a> of the liquid cultures in a 96-well plate of a TECAN reader and can be described in ordinary differential equations (ODEs). |
| + | Such ODEs contain parameters that characterize e.g. growth or synthesis rates for bacteria or indigoidine, respectively. | ||
</p> | </p> | ||
<h3 id="ODEs">Ordinary Differential Equations (ODEs)</h3> | <h3 id="ODEs">Ordinary Differential Equations (ODEs)</h3> | ||
<p> | <p> | ||
| - | + | How to find proper equations for bacterial growth and indigoidine synthesis? | |
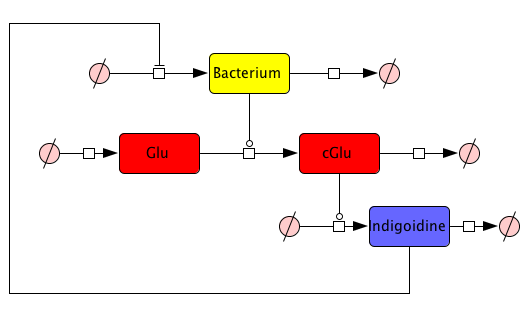
| - | From our mind model (<a class="fancybox fancyFigure" href="/wiki/images/0/03/Network_native.png" title="<b>Figure 2: Schematic representation of a mathematical model describing bacterial Indigoidine synthesis</b> | + | From our mind model (<a class="fancybox fancyFigure" href="/wiki/images/0/03/Network_native.png" title="<b>Figure 2: Schematic representation of a mathematical model describing bacterial Indigoidine synthesis</b>.">Fig. 2</a>), we derived ODEs based on mass-action kinetics[2]. However, bacterial growth curves cannot be sufficiently described by mass action, thus we adapted our ODE for the bacterial growth from equation (7) of Kenneth and Kamau, 1993[3]. |
Our ODE system is now given by the following 4 equations: | Our ODE system is now given by the following 4 equations: | ||
</p> | </p> | ||
<p> | <p> | ||
\begin{align} | \begin{align} | ||
| - | \mathrm{d}\mathrm{[Bac]}/\mathrm{d}t &= \frac{\mathrm{[Bac]} \cdot \left(\mathrm{ | + | \mathrm{d}\mathrm{[Bac]}/\mathrm{d}t &= \frac{\mathrm{[Bac]} \cdot \left(\mathrm{Bacmax} - \mathrm{[Bac]}\right) \cdot \mathrm{beta}}{\mathrm{Bacmax}}\label{bacgrowth}\\ |
\mathrm{d}\mathrm{[Glu]}/\mathrm{d}t &= - \mathrm{[Bac]} \cdot \mathrm{[Glu]} \cdot \mathrm{ksyn}\label{glu}\\ | \mathrm{d}\mathrm{[Glu]}/\mathrm{d}t &= - \mathrm{[Bac]} \cdot \mathrm{[Glu]} \cdot \mathrm{ksyn}\label{glu}\\ | ||
| Line 40: | Line 50: | ||
Initially, inhibition of bacterial growth by indigoidine and degradation of cyclic glutamine were also described by the model, using equations \eqref{bacgrowthinhib} and \eqref{cgludeg} instead of \eqref{bacgrowth} and \eqref{cglu}, respectively, however the degradation rate turned out to be non-identifiable for all data sets, converging to the lower bound, whereas the growth inhibition term led to frequent convergence failures, the inhibition constant being very low in cases where convergence was achieved. These two terms were thus removed from the model. | Initially, inhibition of bacterial growth by indigoidine and degradation of cyclic glutamine were also described by the model, using equations \eqref{bacgrowthinhib} and \eqref{cgludeg} instead of \eqref{bacgrowth} and \eqref{cglu}, respectively, however the degradation rate turned out to be non-identifiable for all data sets, converging to the lower bound, whereas the growth inhibition term led to frequent convergence failures, the inhibition constant being very low in cases where convergence was achieved. These two terms were thus removed from the model. | ||
\begin{align} | \begin{align} | ||
| - | \mathrm{d}\mathrm{[Bac]}/\mathrm{d}t &= \frac{\mathrm{[Bac]} \cdot \left(\mathrm{ | + | \mathrm{d}\mathrm{[Bac]}/\mathrm{d}t &= \frac{\mathrm{[Bac]} \cdot \left(\mathrm{Bacmax} - \mathrm{[Bac]}\right) \cdot \left(\mathrm{beta} - \mathrm{ki} \cdot \mathrm{[Ind]}\right)}{\mathrm{Bacmax}}\label{bacgrowthinhib}\\ |
\mathrm{d}\mathrm{[cGlu]}/\mathrm{d}t &= - \mathrm{kdim} \cdot {\mathrm{[cGlu]}}^2 - \mathrm{kdegg} \cdot \mathrm{[cGlu]} + \mathrm{[Bac]} \cdot \mathrm{[Glu]} \cdot \mathrm{ksyn}\label{cgludeg} | \mathrm{d}\mathrm{[cGlu]}/\mathrm{d}t &= - \mathrm{kdim} \cdot {\mathrm{[cGlu]}}^2 - \mathrm{kdegg} \cdot \mathrm{[cGlu]} + \mathrm{[Bac]} \cdot \mathrm{[Glu]} \cdot \mathrm{ksyn}\label{cgludeg} | ||
| Line 48: | Line 58: | ||
This system contains 4 dynamic variables: Bacteria (Bac), Glutamine (Glu), cyclized Glutamine (cGlu) and Indigoidine (Ind) that change with time t. | This system contains 4 dynamic variables: Bacteria (Bac), Glutamine (Glu), cyclized Glutamine (cGlu) and Indigoidine (Ind) that change with time t. | ||
Bacteria and Indigoidine was experimentally measured, we thus call Bac and Ind observables of our system. | Bacteria and Indigoidine was experimentally measured, we thus call Bac and Ind observables of our system. | ||
| - | The equations are described by 5 kinetic parameters: | + | The equations are described by 5 kinetic parameters:</p> |
<ul> | <ul> | ||
<li> Bacmax: maximum capacity for bacterial growth | <li> Bacmax: maximum capacity for bacterial growth | ||
| Line 56: | Line 66: | ||
<li> kdegi: degradation rate of indigoidine | <li> kdegi: degradation rate of indigoidine | ||
</ul> | </ul> | ||
| - | In addition, the experimental error for the observables was estimated with 2 error parameters and the initial concentration of the bacteria at t=0 was estimated. Data was otherwise normalized between 0 and 1, thus no scaling and offset parameters were required. In order to keep the model simple, it does not contain glutamine production, the initial glutamine concentration was arbitrarily set to 1. While this does not permit conclusions about absolute indigoidine synthesis rates, comparison of rates between the individual conditions is possible. | + | <p>In addition, the experimental error for the observables was estimated with 2 error parameters and the initial concentration of the bacteria at t=0 was estimated. Data was otherwise normalized between 0 and 1, thus no scaling and offset parameters were required. In order to keep the model simple, it does not contain glutamine production, the initial glutamine concentration was arbitrarily set to 1. While this does not permit conclusions about absolute indigoidine synthesis rates, comparison of rates between the individual conditions is possible. |
</p> | </p> | ||
<center> | <center> | ||
| - | <a class="fancybox fancyFigure" href="/wiki/images/0/03/Network_native.png" title="<b>Figure 2: Schematic representation of a mathematical model describing bacterial Indigoidine synthesis</b> | + | <a class="fancybox fancyFigure" href="/wiki/images/0/03/Network_native.png" title="<b>Figure 2: Schematic representation of a mathematical model describing bacterial Indigoidine synthesis</b>."> |
<img style="width:30%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/0/03/Network_native.png"/> | <img style="width:30%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/0/03/Network_native.png"/> | ||
<figcaption style="width:60%;"><b>Figure 2: Schematic representation of a mathematical model describing bacterial Indigoidine synthesis</b>. ....</figcaption> | <figcaption style="width:60%;"><b>Figure 2: Schematic representation of a mathematical model describing bacterial Indigoidine synthesis</b>. ....</figcaption> | ||
| Line 67: | Line 77: | ||
<h3 id="D2D">Framework</h3> | <h3 id="D2D">Framework</h3> | ||
<p> | <p> | ||
| - | Parameters have to be estimated from experimental data. In order to implement our mathematical model and the wetlab data, we used an open-source software package allowing for comprehensive analysis (<a href="https://bitbucket.org/d2d-development/d2d-software/wiki/Home">D2D Software</a>). With this framework, we were able to calibrate the model and perform robust parameter estimations | + | Parameters have to be estimated from experimental data. In order to implement our mathematical model and the wetlab data, we used an open-source software package allowing for comprehensive analysis (<a href="https://bitbucket.org/d2d-development/d2d-software/wiki/Home">D2D Software</a>). With this framework, we were able to calibrate the model and perform robust parameter estimations[4] as well as identifiability analysis[5] yielding valuable insight into bacterial indigoidine production. |
</p> | </p> | ||
<h2 id="Results">Results</h2> | <h2 id="Results">Results</h2> | ||
<p> | <p> | ||
| - | The data sets could be grouped into two types, those with a significant indigoidine production and those without. Although both types of data sets resulted in seemingly good fits (Fig. 4), identifiability analysis using a profile likelihood exploiting approach | + | We received experimental data from the indigoidine team, who wanted to compare indigoidine synthesis rates between various T domains and PPTases. The model described above was thus fitted individually to each T domain / PPTase combination and simultaneously to all replicates. The data sets could be grouped into two types, those with a significant indigoidine production and those without. Although both types of data sets resulted in seemingly good fits (Fig. 4), identifiability analysis using a profile likelihood exploiting approach[6] showed that for all data sets without indigoidine production at least one parameter could not be reliably estimated (an exemplary plot is shown in Fig. 5B), whereas all parameters were identifiable for data sets showing indigoidine production (Fig. 5A). As our model only permits relative quantification of synthesis rates due to the lack of glutamine production and the arbitrarily chosen initial glutamine concentration, we normalized the indigoidine synthesis rates of the fully identifiable data sets to the range [0, 1] and visualized them in a heatmap (Fig. 3). SynT1 seems to be the most effective T domain for the entD and sfp PPTases, which corresponds very well to the preliminary analysis performed by the indigoidine team. |
</p> | </p> | ||
<center> | <center> | ||
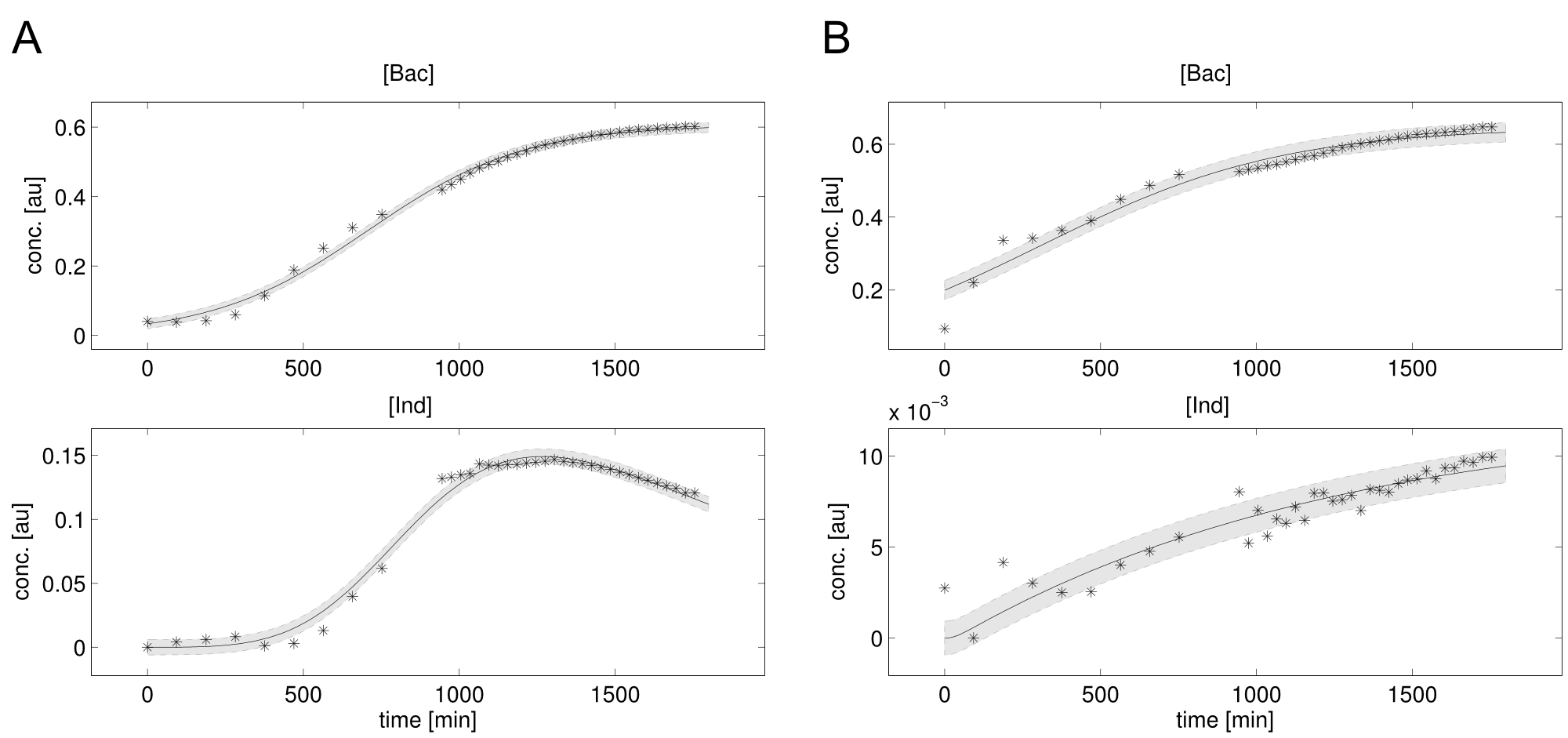
| - | <a class="fancybox fancyFigure" href="/wiki/images/4/45/Heidelberg_fit-native_sfp-T10_delC.png" title="<b>Figure 3: Experimental data (stars) and simulation (lines) of bacterial growth and indigoidine concentration.</b> Shadings denote the 95% confidence interval. <b>A)</b> Native indC with the sfp PPTase. <b>B)</b> | + | <a class="fancybox fancyFigure" href="/wiki/images/4/45/Heidelberg_fit-native_sfp-T10_delC.png" title="<b>Figure 3: Experimental data (stars) and simulation (lines) of bacterial growth and indigoidine concentration.</b> Shadings denote the 95% confidence interval. <b>A)</b> Native indC with the sfp PPTase. <b>B)</b> SynT1 with the delC PPTase."> |
<img style="width:100%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/4/45/Heidelberg_fit-native_sfp-T10_delC.png"/> | <img style="width:100%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/4/45/Heidelberg_fit-native_sfp-T10_delC.png"/> | ||
| - | <figcaption style="width:100%"><b>Figure 3: Experimental data (stars) and simulation (lines) of bacterial growth and indigoidine concentration.</b> Shadings denote the 95% confidence interval. <b>A)</b> Native indC with the sfp PPTase. <b>B)</b> | + | <figcaption style="width:100%"><b>Figure 3: Experimental data (stars) and simulation (lines) of bacterial growth and indigoidine concentration.</b> Shadings denote the 95% confidence interval. <b>A)</b> Native indC with the sfp PPTase. <b>B)</b> SynT1 with the delC PPTase.</figcaption> |
</a> | </a> | ||
</center> | </center> | ||
<center> | <center> | ||
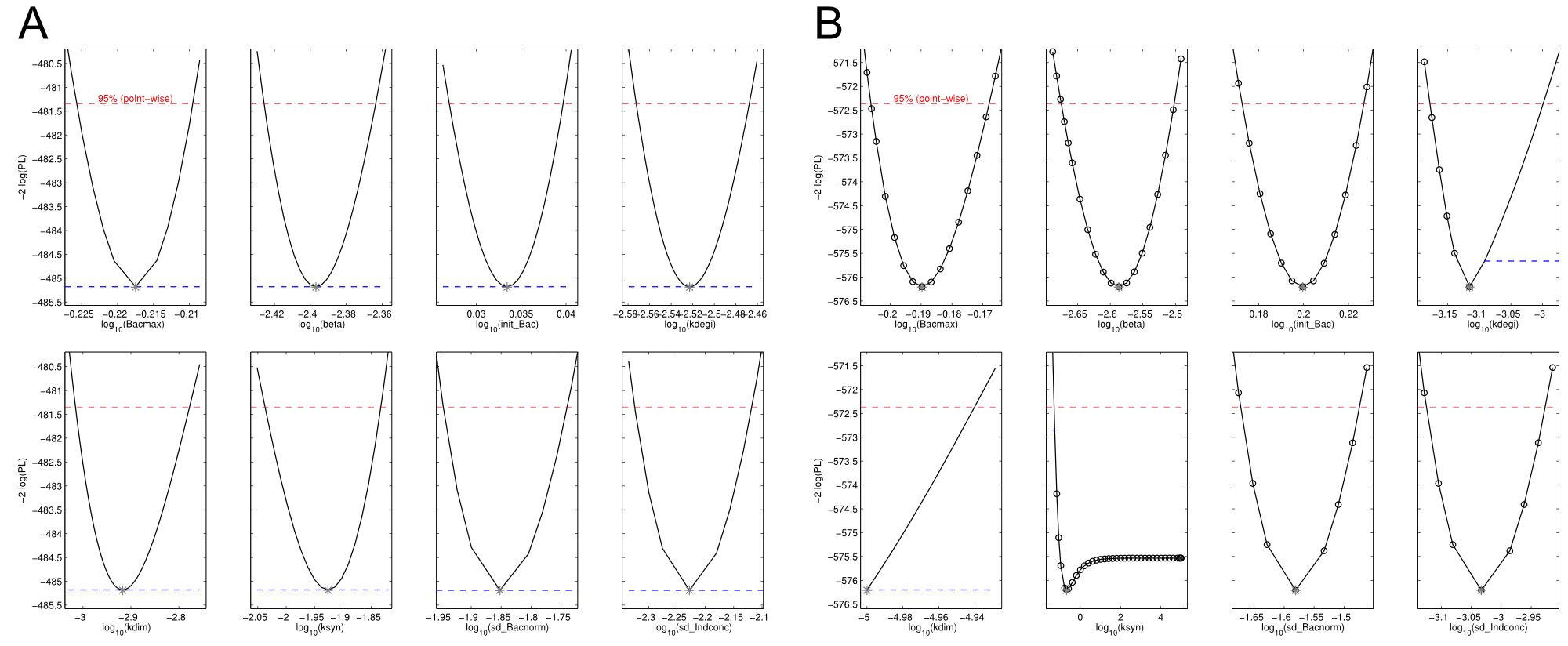
| - | <a class="fancybox fancyFigure" href="/wiki/images/9/94/Heidelberg_ple-native_sfp-T10_delC.png" title="<b>Figure 4: Identifiability analysis of the fitted parameters was performed using a profile likelihood expoiting approach.</b> <b>A)</b> Native indC with the sfp PPTase. All parameters are fully identifiable. <b>B)</b> | + | <a class="fancybox fancyFigure" href="/wiki/images/9/94/Heidelberg_ple-native_sfp-T10_delC.png" title="<b>Figure 4: Identifiability analysis of the fitted parameters was performed using a profile likelihood expoiting approach.</b> <b>A)</b> Native indC with the sfp PPTase. All parameters are fully identifiable. <b>B)</b> SynT1 with the delC PPTase. kdim and ksyn are not identifiable with ksyn converging to the lower bound."> |
<img style="width:100%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/9/94/Heidelberg_ple-native_sfp-T10_delC.png"/> | <img style="width:100%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/9/94/Heidelberg_ple-native_sfp-T10_delC.png"/> | ||
| - | <figcaption style="width:100%"><b>Figure 4: Identifiability analysis of the fitted parameters was performed using a profile likelihood expoiting approach.</b> <b>A)</b> Native indC with the sfp PPTase. All parameters are fully identifiable. <b>B)</b> | + | <figcaption style="width:100%"><b>Figure 4: Identifiability analysis of the fitted parameters was performed using a profile likelihood expoiting approach.</b> <b>A)</b> Native indC with the sfp PPTase. All parameters are fully identifiable. <b>B)</b> SynT1 with the delC PPTase. kdim and ksyn are not identifiable with ksyn converging to the lower bound.</figcaption> |
</a> | </a> | ||
</center> | </center> | ||
<center> | <center> | ||
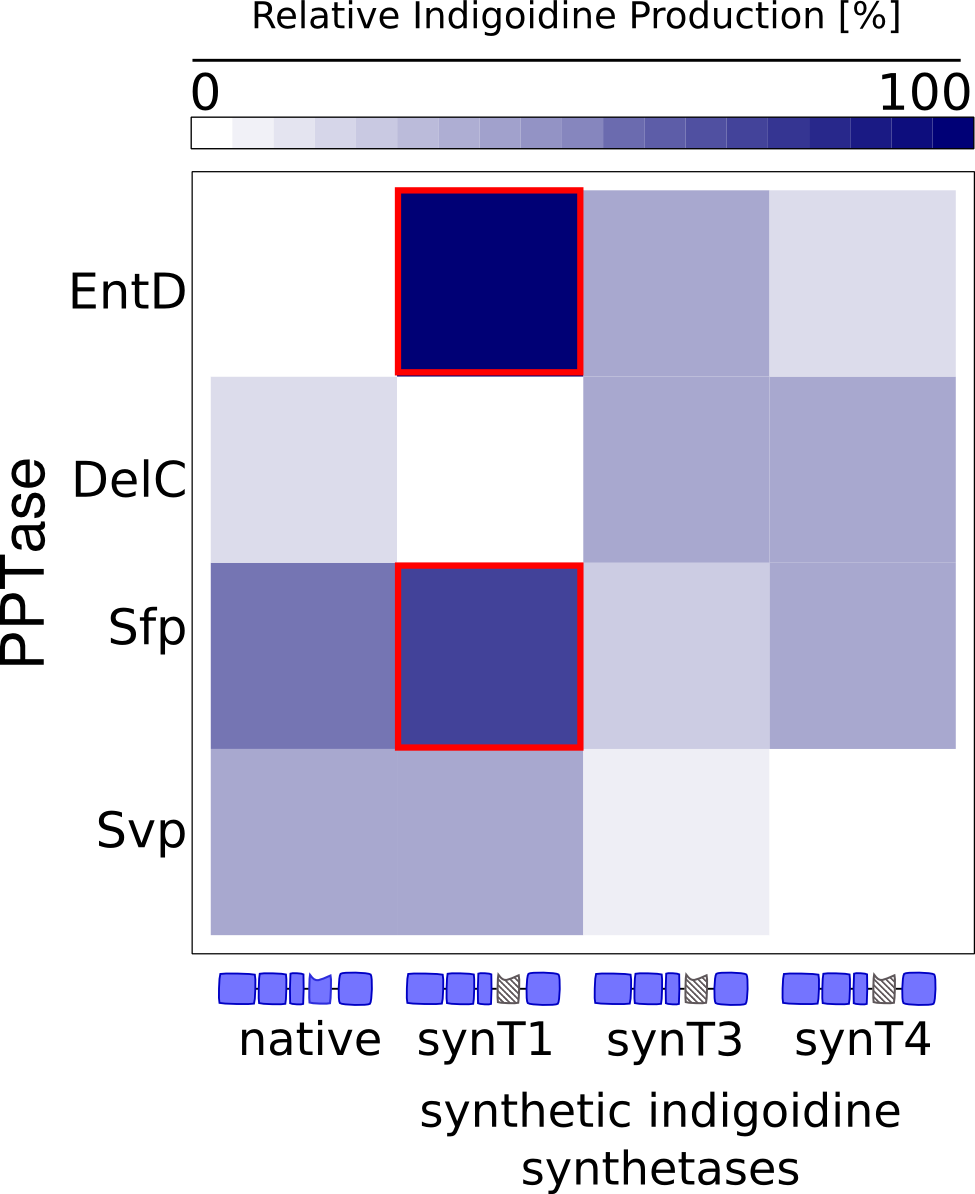
| - | <a class="fancybox fancyFigure" href="/wiki/images/5/55/Heidelberg_heatmap_model1.png" title="<b>Figure 5: Indigoidine synthesis rates ksyn determined by fitting the model to several data sets</b>. The rates were normalized to the range between 0 and 1, rates for data sets where at least one parameter was not identifiable were set to 0. | + | <a class="fancybox fancyFigure" href="/wiki/images/5/55/Heidelberg_heatmap_model1.png" title="<b>Figure 5: Indigoidine synthesis rates ksyn determined by fitting the model to several data sets</b>. The rates were normalized to the range between 0 and 1, rates for data sets where at least one parameter was not identifiable were set to 0. SynT1 is the most efficient of all tested T domains, whereas synT3 is the only one that functions with all PPTases."> |
<img style="width:30%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/5/55/Heidelberg_heatmap_model1.png"/> | <img style="width:30%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/5/55/Heidelberg_heatmap_model1.png"/> | ||
| - | <figcaption style="width:60%;"><b>Figure 5: Indigoidine synthesis rates ksyn determined by fitting the model to several data sets</b>. The rates were normalized to the range between 0 and 1, rates for data sets where at least one parameter was not identifiable were set to 0. The synthetic T domain | + | <figcaption style="width:60%;"><b>Figure 5: Indigoidine synthesis rates ksyn determined by fitting the model to several data sets</b>. The rates were normalized to the range between 0 and 1, rates for data sets where at least one parameter was not identifiable were set to 0. The synthetic T domain synT1 is the most efficient of all tested T domains, whereas synT3 is the only one that functions with all PPTases.</figcaption> |
</a> | </a> | ||
</center> | </center> | ||
| Line 97: | Line 107: | ||
Our mathematical model was able to describe most of our experimental data and was in those cases structurally and practically fully identifiable. We thus challenged the model to deduce additional facts on indigoidine synthesis. | Our mathematical model was able to describe most of our experimental data and was in those cases structurally and practically fully identifiable. We thus challenged the model to deduce additional facts on indigoidine synthesis. | ||
| - | First, we had a look at the parameters that were estimated to describe our experimental data sets. For each parameter estimate, the lower boundary was set to 1e-5 and the upper boundary was set to 1e+5. Initial guesses for parameters during parameter estimations were generated via latin hypercube sampling to ensure broad coverage of all regions in the high-dimensional parameter space | + | First, we had a look at the parameters that were estimated to describe our experimental data sets. For each parameter estimate, the lower boundary was set to 1e-5 and the upper boundary was set to 1e+5. Initial guesses for parameters during parameter estimations were generated via latin hypercube sampling to ensure broad coverage of all regions in the high-dimensional parameter space[7]. Nevertheless, some parameters exhibited broad variation among the best fits of the different data sets while other parameters were estimated in a narrow range (<a class="fancybox fancyFigure" href="/wiki/images/d/d9/Heidelberg_boxplot1.png" title="<b>Figure 6: Box and whiskers plot of the parameters fitted for the invidual data sets.</b> The black line represents the median, the box ranges from the first to the third quartile of the sample. The dashed whiskers denote the median ± 1.5 times the interquartile range. The red line denotes the mean, yellow lines the mean ± standard error of the mean, solid whiskers the mean ± standard deviation.">Fig. 6</a>). |
</p> | </p> | ||
<center> | <center> | ||
| - | <a class="fancybox fancyFigure" href="/wiki/images/d/d9/Heidelberg_boxplot1.png" title="<b>Figure | + | <a class="fancybox fancyFigure" href="/wiki/images/d/d9/Heidelberg_boxplot1.png" title="<b>Figure 6: Box and whiskers plot of the parameters fitted for the invidual data sets.</b> The black line represents the median, the box ranges from the first to the third quartile of the sample. The dashed whiskers denote the median ± 1.5 times the interquartile range. The red line denotes the mean, yellow lines the mean ± standard error of the mean, solid whiskers the mean ± standard deviation."> |
<img style="width:30%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/d/d9/Heidelberg_boxplot1.png"/> | <img style="width:30%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/d/d9/Heidelberg_boxplot1.png"/> | ||
| - | <figcaption style="width:60%"><b>Figure | + | <figcaption style="width:60%"><b>Figure 6: Box and whiskers plot of the parameters fitted for the invidual data sets.</b> The black line represents the median, the box ranges from the first to the third quartile of the sample. The dashed whiskers denote the median ± 1.5 times the interquartile range. The red line denotes the mean, yellow lines the mean ± standard error of the mean, solid whiskers the mean ± standard deviation.</figcaption> |
</a> | </a> | ||
</center> | </center> | ||
<p> | <p> | ||
Interestingly, the bacterial growth rate beta and the maximum growth capacity Bacmax were very consistent. | Interestingly, the bacterial growth rate beta and the maximum growth capacity Bacmax were very consistent. | ||
| - | The parameters ksyn, kdim and kdegi contributed directly to indogoidine synthesis as it can be inferred from the above mentioned equations | + | The parameters ksyn, kdim and kdegi contributed directly to indogoidine synthesis as it can be inferred from the above mentioned equations \eqref{glu}, \eqref{cglu} and \eqref{ind}. |
The synthesis rate ksyn characterizes the cyclization of glutamine. The dimerization rate kdim reflects how two cyclized glutamine molecules form one indgoidine molecule. As such, ksyn and kdim represent the efficiency of the T domain of indC. | The synthesis rate ksyn characterizes the cyclization of glutamine. The dimerization rate kdim reflects how two cyclized glutamine molecules form one indgoidine molecule. As such, ksyn and kdim represent the efficiency of the T domain of indC. | ||
The degradation rate of indigoidine, kdegi, should be independent from the domain structure of the indigoidine synthetase as the stability of the peptide is not affected by the T domains or the PPTases that were tested. The rate did not vary widely and the diverse data sets could still be explained by the mathematical model. At first glance, kdegi seemed less important for our experimentally assessed conditions. To further investigate the role of kdegi for indigoidine production kinetics, we systematically varied this parameter and compared simulation results. | The degradation rate of indigoidine, kdegi, should be independent from the domain structure of the indigoidine synthetase as the stability of the peptide is not affected by the T domains or the PPTases that were tested. The rate did not vary widely and the diverse data sets could still be explained by the mathematical model. At first glance, kdegi seemed less important for our experimentally assessed conditions. To further investigate the role of kdegi for indigoidine production kinetics, we systematically varied this parameter and compared simulation results. | ||
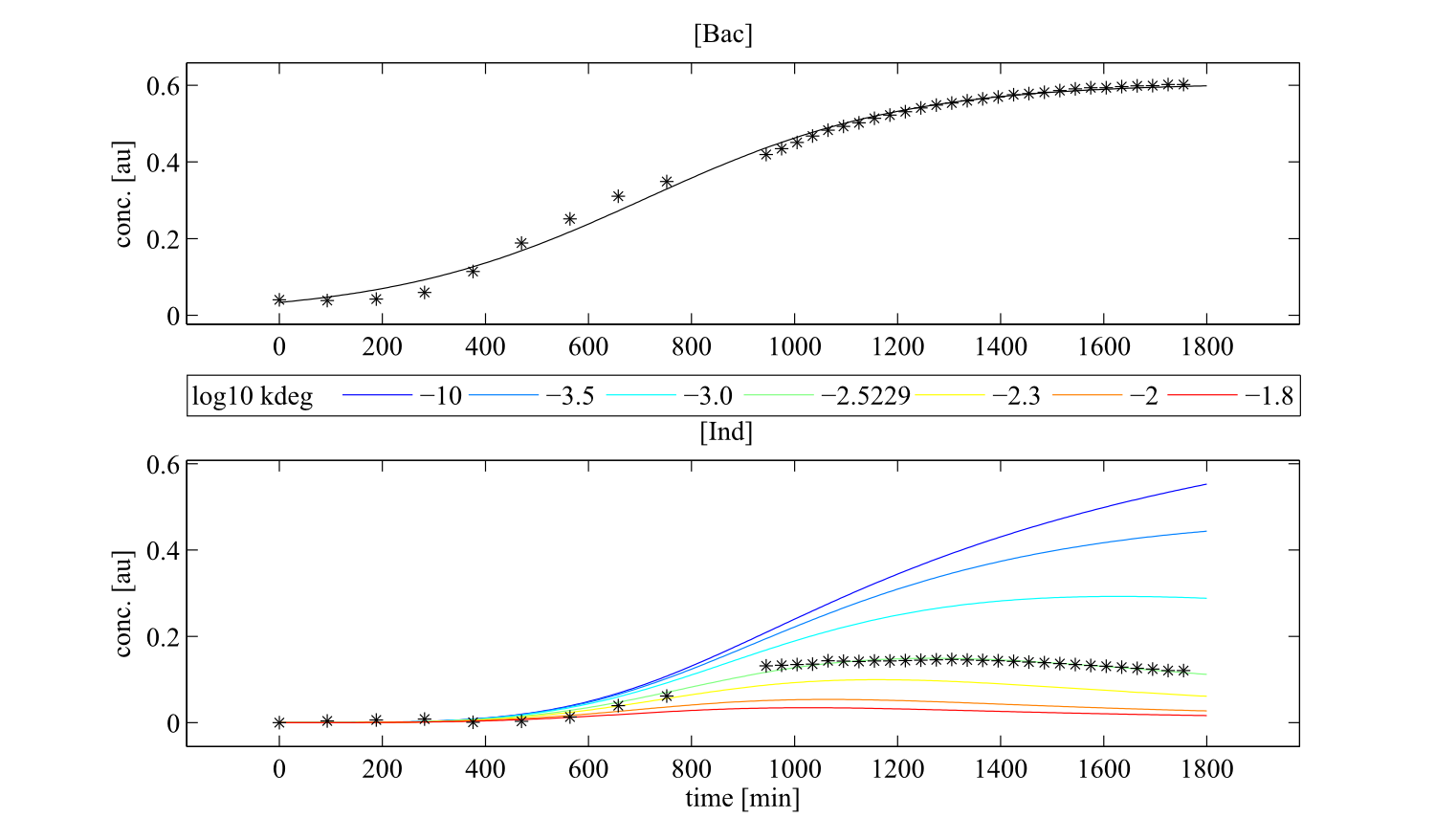
| - | The synthesis of indigoidine was clearly affected by variation of kdegi (Fig. | + | The synthesis of indigoidine was clearly affected by variation of kdegi (<a class="fancybox fancyFigure" href="/wiki/images/7/7b/Heidelberg_kdeg_variation_nsfp.png" title="<b>Figure 7: Dependency of indigoidine production on the degradation constant.</b>">Fig. 7</a>). The [Ind] trajectory collapsed more than 3-fold increase of the degradation rate. On the contrary, improved peptide stability could yield much more indigoidine. For very low degradation rates (i.e. kdegi < 1e-3), synthesis would not even saturate within the observed time frame. |
</p> | </p> | ||
<center> | <center> | ||
| - | <a class="fancybox fancyFigure" href="/wiki/images/7/7b/Heidelberg_kdeg_variation_nsfp.png" title="<b>Figure | + | <a class="fancybox fancyFigure" href="/wiki/images/7/7b/Heidelberg_kdeg_variation_nsfp.png" title="<b>Figure 7: Dependency of indigoidine production on the degradation constant.</b> The analysis was performed for the native indC with the sfp PPTase as a representative example."> |
<img style="width:60%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/7/7b/Heidelberg_kdeg_variation_nsfp.png"/> | <img style="width:60%; margin-bottom:10px; margin-top:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="/wiki/images/7/7b/Heidelberg_kdeg_variation_nsfp.png"/> | ||
| - | <figcaption style="width:60%"><b>Figure | + | <figcaption style="width:60%"><b>Figure 7: Dependency of indigoidine production on the degradation constant.</b> The analysis was performed for the native indC with the sfp PPTase as a representative example.</figcaption> |
</a> | </a> | ||
</center> | </center> | ||
| Line 121: | Line 131: | ||
<h3 id="SensitivityAnalysis"> Sensitivity analysis for optimized indigoidine production </h3> | <h3 id="SensitivityAnalysis"> Sensitivity analysis for optimized indigoidine production </h3> | ||
<p> | <p> | ||
| - | To perceive the role of kdegi for indigoidine synthesis, we have varied the parameter over a broad range and observed qualitatively distinct model trajectories. For a more quantitative understanding of how indigoidine yield changes with altering parameters of the system, we conducted a sensitivity analysis | + | To perceive the role of kdegi for indigoidine synthesis, we have varied the parameter over a broad range and observed <a class="fancybox fancyFigure" href="/wiki/images/7/7b/Heidelberg_kdeg_variation_nsfp.png" title="<b>Figure 7: Dependency of indigoidine production on the degradation constant.</b>">qualitatively distinct model trajectories</a>. For a more quantitative understanding of how indigoidine yield changes with altering parameters of the system, we conducted a sensitivity analysis[8]. |
| - | Therefore, we first defined the variable IntInd, the integrated indigoidine reflecting production yield. The change of the indigoidine concentration with time is determined by the ODE | + | Therefore, we first defined the variable IntInd, the integrated indigoidine reflecting production yield. The change of the indigoidine concentration with time is determined by the ODE \eqref{ind} and its integral with time can be depicted as the area under the [Ind] curve, i.e. the amount of indigoidine produced by the bacteria with time. Now we were interested in the change of IntInd with the change of the different parameters of our system. |
| - | Mathematically, this is expressed as the so called sensitivity $\frac{\partial \mathrm{X}}{\partial \mathrm{p}}, with X as our variable of interest (IntInd) and p as the parameter (beta, Bacmax, ksyn, kdim or kdegi, respectively). This partial derivative provides a measure of how much X changes with the change of p or how sensitive X is against small variations in p. Sensitivities can be scaled with the actual values of X and the respective p to fall in the range from -1 to +1. The higher the absolute value of the sensitivity, the more important is the parameter for the variable. A positive sensitivity means positive impact of the parameter on the variable whereas a sensitivity < 0 indicates reduction of X for subtle decrease in p. | + | Mathematically, this is expressed as the so called sensitivity $\frac{\partial \mathrm{X}}{\partial \mathrm{p}}$, with X as our variable of interest (IntInd) and p as the parameter (beta, Bacmax, ksyn, kdim or kdegi, respectively). This partial derivative provides a measure of how much X changes with the change of p or how sensitive X is against small variations in p. Sensitivities can be scaled with the actual values of X and the respective p to fall in the range from -1 to +1. The higher the absolute value of the sensitivity, the more important is the parameter for the variable. A positive sensitivity means positive impact of the parameter on the variable whereas a sensitivity < 0 indicates reduction of X for subtle decrease in p. |
We utilised our mathematical model and computed the sensitivities of IntInd against the change of all five kinetic parameters of our system and scaled them accordingly. The analysis confirmed that the kdegi was the only parameter of the system that exhibited a negative impact on the indigoidine production (Tab. 1). Slight increase of all the other parameters also augmented additional yield. | We utilised our mathematical model and computed the sensitivities of IntInd against the change of all five kinetic parameters of our system and scaled them accordingly. The analysis confirmed that the kdegi was the only parameter of the system that exhibited a negative impact on the indigoidine production (Tab. 1). Slight increase of all the other parameters also augmented additional yield. | ||
</p> | </p> | ||
| Line 137: | Line 147: | ||
<p> | <p> | ||
The initial concentration of bacteria represented the most crucial parameter for indigoidine production, which was intuitive since the amount of produced indigoidine depends on the amount of bacteria in culture that synthesise the peptide. | The initial concentration of bacteria represented the most crucial parameter for indigoidine production, which was intuitive since the amount of produced indigoidine depends on the amount of bacteria in culture that synthesise the peptide. | ||
| - | The parameter kdegi exhibited a high impact on IntInd but did not vary so much among the experimental conditions. One would assume that the half-life of the peptide is not affected by different T domains or PPTases. Possibly, the arrangement of modules influences peptide stability as inter modular communication is a precondition for robust peptide assembly | + | The parameter kdegi exhibited a high impact on IntInd but did not vary so much among the experimental conditions. One would assume that the half-life of the peptide is not affected by different T domains or PPTases. Possibly, the arrangement of modules influences peptide stability as inter-modular communication is a precondition for robust peptide assembly[9]. This links the peptide synthesis optimisation to our <a href="https://2013.igem.org/Team:Heidelberg/Project/Tyrocidine">module shuffling project</a> where modules of different hosts were combined for custom peptide synthesis. |
Indigoidine yield was also sensitive against small variations in beta and Bacmax. Culture conditions could be improved and metabolically more active strains could be chosen to further optimize performance in this respect. | Indigoidine yield was also sensitive against small variations in beta and Bacmax. Culture conditions could be improved and metabolically more active strains could be chosen to further optimize performance in this respect. | ||
| - | The parameters ksyn and kdim also determine peptide production. | + | The parameters ksyn and kdim also determine peptide production. |
</p> | </p> | ||
<p> | <p> | ||
| - | The question is whether those rates could be altered by engineering the NRPS to advanced performance. To answer this question, we performed | + | The question is whether those rates could be altered by engineering the NRPS to advanced performance. To answer this question, we performed multiple linear regressions. We regressed the alignment score of the synthetic T domains of the indigoidine synthase with the rates from the respective best fit. The score was calculated <a href="http://www.ebi.ac.uk/Tools/msa/clustalw2/">by pairwise multiple sequence alignment</a> of the amino acid sequences of the synthetic T domains with the native T domain of the indigoidine synthase as a template. The parameter values were obtained from parameter estimations of our mathematical model that has been calibrated with experimental data as described above. Linear regression of the alignment score on ksyn did only result in a coefficient of correlation of 0.48 while kdim correlated rather well (R<sup>2</sup>=0.97). This pinpointed the possibility to engineer T domains of NRPSs to optimize kinetic rates <i>in silico</i> for peptide production. |
</p> | </p> | ||
<h2 id="Conclusion"> Conclusion and Outlook </h2> | <h2 id="Conclusion"> Conclusion and Outlook </h2> | ||
| - | + | <p> | |
| - | indigoidine to | + | We set up a mathematical model for bacterial indigoidine production to showcase the systematic scrutinization of NRP synthesis kinetics. |
| - | + | In the course of model calibration with experimental data we could already falsify the hypothesis that indigoidine had an inhibitory effect on growth of the bacteria. The model with such inhibitory terms was not able to describe the experimental data. | |
| + | A reduced model was proven to be fully identifiable via a profile likelihood approach and simulations could reproduce experimental findings. | ||
| + | The impact of the different rates on indigoidine production was investigated via sensitivity analysis. | ||
| + | We can conclude that yield could be increased when peptide stability is improved and synthesis efficiency is advanced. Linear regression suggests that the dimerization rate of indigoidine could be engineered by editing the T domain sequence. | ||
| + | With this, we have learnt valuable lessons on optimizing production of synthetic peptides <i>in silico</i>. Of course, dependency of yield on kinetic parameters does not necessarily scale linearly with peptide length but with these initial trials the way is paved for further analysis. | ||
| + | </p> | ||
| - | </div></div></div> | + | </div> |
| + | <div class="col-sm-12 jumbotron"> | ||
| + | <div class="references"> | ||
| + | <p>1. Brachmann, Alexander O, Kirchner, Ferdinand, Kegler, Carsten, Kinski, Sebastian C, Schmitt, Imke, Bode, Helge B: Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens., J. Biotechnol. 157(1), 96–9, January 2012</p> | ||
| + | <p>2. Klipp, Edda, Liebermeister, Wolfram: Mathematical modeling of intracellular signaling pathways., BMC Neurosci 7 Suppl 1, S10, 2006</p> | ||
| + | <p>3. Pruitt, Kenneth M., Kamau, David N.: Mathematical models of bacterial growth, inhibition and death under combined stress conditions, Journal of Industrial Microbiology 12(3-5), Springer, 221, 1993</p> | ||
| + | <p>4. Raue, Andreas, Schilling, Marcel, Bachmann, Julie, Matteson, Andrew, Schelke, Max, Kaschek, Daniel, Hug, Sabine, Kreutz, Clemens, Harms, Brian D, Theis, Fabian J, Klingmüller, Ursula, Timmer, Jens: Lessons learned from quantitative dynamical modeling in systems biology., PLoS ONE 8(9), e74335, 2013</p> | ||
| + | <p>5. Raue, A, Becker, V, Klingmüller, U, Timmer, J: Identifiability and observability analysis for experimental design in nonlinear dynamical models., Chaos 20(4), 045105, December 2010</p> | ||
| + | <p>6. Raue, A, Kreutz, C, Maiwald, T, Bachmann, J, Schilling, M, Klingmüller, U, Timmer, J: Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood., Bioinformatics 25(15), 1923–9, August 2009</p> | ||
| + | <p>7. Owen, Art B.: A Central Limit Theorem for Latin Hypercube Sampling, Journal of the Royal Statistical Society. Series B (Methodological) 54(2), Wiley for the Royal Statistical Society, 541–551, 1992</p> | ||
| + | <p>8. Zi, Zhike, Zheng, Yanan, Rundell, Ann E, Klipp, Edda: SBML-SAT: a systems biology markup language (SBML) based sensitivity analysis tool., BMC Bioinformatics 9, 342, 2008</p> | ||
| + | <p>9. Gokhale, R S, Tsuji, S Y, Cane, D E, Khosla, C: Dissecting and exploiting intermodular communication in polyketide synthases., Science 284(5413), 482–5, April 1999</p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
</html> | </html> | ||
Latest revision as of 14:27, 30 November 2013
Challenge
A challenge we had to face during the characterization and optimization of indC was to identify the production kinetics of indigoidine. In order to disentangle the underlying mechanisms of bacterial growth and peptide synthesis, we decided to set up a mathematical model based on coupled ordinary differential equations (ODEs). Calibrated with our experimental time-resolved data, the mathematical model could potentially not only elucidate how indigoidine production influences growth of bacteria but also provide a more quantitative understanding of the synthesis efficiency of the different T domains and PPTases that were tested.Approach
First, we set up a mind model based on the fact that the functional indigoidine dimer is produced from two glutamines (Glu) that are each cyclized (cGlu)[1] (Fig. 1), and our observation that indigoidine (Ind)-producing bacteria (Bac) grow slower than mock controls. Those hypotheses resulted in a general model scheme depicting the interdependency between indigoidine synthesis and bacterial growth. With the mathematical model, we could then validate whether there is indeed a negative feedback from the indigoidine production to the growth of bacteria.
Since we had already established our quantitative indigoidine production assay (see Tag-Optimization) in a time-dependent manner, we wanted to further exploit these experimental data via quantitative dynamic modeling. The change of bacteria and Indigoidine with time was measured via optical density of the liquid cultures in a 96-well plate of a TECAN reader and can be described in ordinary differential equations (ODEs). Such ODEs contain parameters that characterize e.g. growth or synthesis rates for bacteria or indigoidine, respectively.
Ordinary Differential Equations (ODEs)
How to find proper equations for bacterial growth and indigoidine synthesis? From our mind model (Fig. 2), we derived ODEs based on mass-action kinetics[2]. However, bacterial growth curves cannot be sufficiently described by mass action, thus we adapted our ODE for the bacterial growth from equation (7) of Kenneth and Kamau, 1993[3]. Our ODE system is now given by the following 4 equations:
\begin{align} \mathrm{d}\mathrm{[Bac]}/\mathrm{d}t &= \frac{\mathrm{[Bac]} \cdot \left(\mathrm{Bacmax} - \mathrm{[Bac]}\right) \cdot \mathrm{beta}}{\mathrm{Bacmax}}\label{bacgrowth}\\ \mathrm{d}\mathrm{[Glu]}/\mathrm{d}t &= - \mathrm{[Bac]} \cdot \mathrm{[Glu]} \cdot \mathrm{ksyn}\label{glu}\\ \mathrm{d}\mathrm{[cGlu]}/\mathrm{d}t &= - \mathrm{kdim} \cdot {\mathrm{[cGlu]}}^2 + \mathrm{[Bac]} \cdot \mathrm{[Glu]} \cdot \mathrm{ksyn}\label{cglu}\\ \mathrm{d}\mathrm{[Ind]}/\mathrm{d}t &= {\mathrm{[cGlu]}}^2 \cdot \mathrm{kdim} - \mathrm{[Ind]} \cdot \mathrm{kdegi}\label{ind} \end{align}
Initially, inhibition of bacterial growth by indigoidine and degradation of cyclic glutamine were also described by the model, using equations \eqref{bacgrowthinhib} and \eqref{cgludeg} instead of \eqref{bacgrowth} and \eqref{cglu}, respectively, however the degradation rate turned out to be non-identifiable for all data sets, converging to the lower bound, whereas the growth inhibition term led to frequent convergence failures, the inhibition constant being very low in cases where convergence was achieved. These two terms were thus removed from the model. \begin{align} \mathrm{d}\mathrm{[Bac]}/\mathrm{d}t &= \frac{\mathrm{[Bac]} \cdot \left(\mathrm{Bacmax} - \mathrm{[Bac]}\right) \cdot \left(\mathrm{beta} - \mathrm{ki} \cdot \mathrm{[Ind]}\right)}{\mathrm{Bacmax}}\label{bacgrowthinhib}\\ \mathrm{d}\mathrm{[cGlu]}/\mathrm{d}t &= - \mathrm{kdim} \cdot {\mathrm{[cGlu]}}^2 - \mathrm{kdegg} \cdot \mathrm{[cGlu]} + \mathrm{[Bac]} \cdot \mathrm{[Glu]} \cdot \mathrm{ksyn}\label{cgludeg} \end{align}
This system contains 4 dynamic variables: Bacteria (Bac), Glutamine (Glu), cyclized Glutamine (cGlu) and Indigoidine (Ind) that change with time t. Bacteria and Indigoidine was experimentally measured, we thus call Bac and Ind observables of our system. The equations are described by 5 kinetic parameters:
- Bacmax: maximum capacity for bacterial growth
- beta: maximum attainable growth rate
- ksyn: synthesis rate of cyclized glutamine from glutamine
- kdim: dimerization rate of two cyclized glutamines to an Indigoidine dimer
- kdegi: degradation rate of indigoidine
In addition, the experimental error for the observables was estimated with 2 error parameters and the initial concentration of the bacteria at t=0 was estimated. Data was otherwise normalized between 0 and 1, thus no scaling and offset parameters were required. In order to keep the model simple, it does not contain glutamine production, the initial glutamine concentration was arbitrarily set to 1. While this does not permit conclusions about absolute indigoidine synthesis rates, comparison of rates between the individual conditions is possible.
Framework
Parameters have to be estimated from experimental data. In order to implement our mathematical model and the wetlab data, we used an open-source software package allowing for comprehensive analysis (D2D Software). With this framework, we were able to calibrate the model and perform robust parameter estimations[4] as well as identifiability analysis[5] yielding valuable insight into bacterial indigoidine production.
Results
We received experimental data from the indigoidine team, who wanted to compare indigoidine synthesis rates between various T domains and PPTases. The model described above was thus fitted individually to each T domain / PPTase combination and simultaneously to all replicates. The data sets could be grouped into two types, those with a significant indigoidine production and those without. Although both types of data sets resulted in seemingly good fits (Fig. 4), identifiability analysis using a profile likelihood exploiting approach[6] showed that for all data sets without indigoidine production at least one parameter could not be reliably estimated (an exemplary plot is shown in Fig. 5B), whereas all parameters were identifiable for data sets showing indigoidine production (Fig. 5A). As our model only permits relative quantification of synthesis rates due to the lack of glutamine production and the arbitrarily chosen initial glutamine concentration, we normalized the indigoidine synthesis rates of the fully identifiable data sets to the range [0, 1] and visualized them in a heatmap (Fig. 3). SynT1 seems to be the most effective T domain for the entD and sfp PPTases, which corresponds very well to the preliminary analysis performed by the indigoidine team.
Our mathematical model was able to describe most of our experimental data and was in those cases structurally and practically fully identifiable. We thus challenged the model to deduce additional facts on indigoidine synthesis. First, we had a look at the parameters that were estimated to describe our experimental data sets. For each parameter estimate, the lower boundary was set to 1e-5 and the upper boundary was set to 1e+5. Initial guesses for parameters during parameter estimations were generated via latin hypercube sampling to ensure broad coverage of all regions in the high-dimensional parameter space[7]. Nevertheless, some parameters exhibited broad variation among the best fits of the different data sets while other parameters were estimated in a narrow range (Fig. 6).
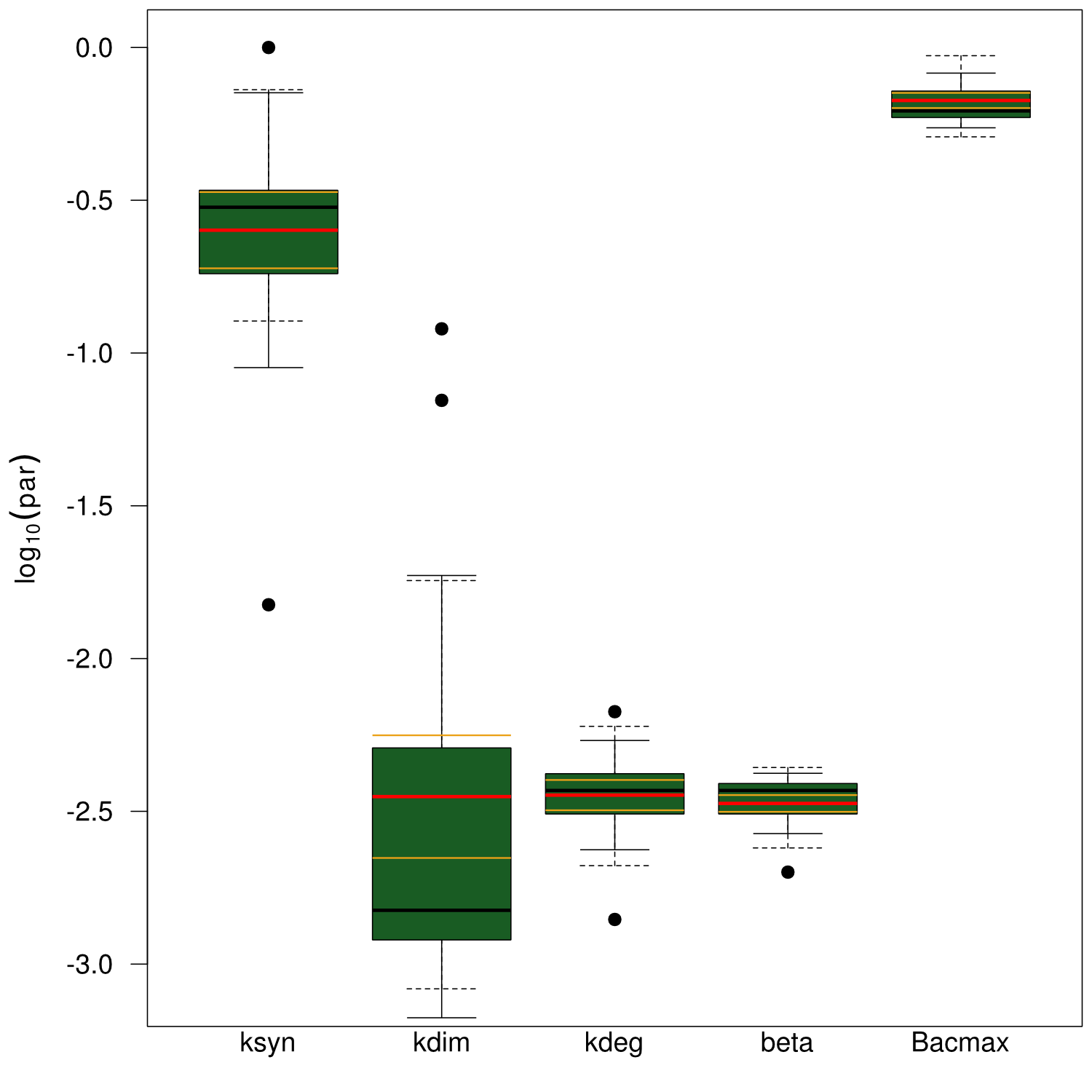
Interestingly, the bacterial growth rate beta and the maximum growth capacity Bacmax were very consistent. The parameters ksyn, kdim and kdegi contributed directly to indogoidine synthesis as it can be inferred from the above mentioned equations \eqref{glu}, \eqref{cglu} and \eqref{ind}. The synthesis rate ksyn characterizes the cyclization of glutamine. The dimerization rate kdim reflects how two cyclized glutamine molecules form one indgoidine molecule. As such, ksyn and kdim represent the efficiency of the T domain of indC. The degradation rate of indigoidine, kdegi, should be independent from the domain structure of the indigoidine synthetase as the stability of the peptide is not affected by the T domains or the PPTases that were tested. The rate did not vary widely and the diverse data sets could still be explained by the mathematical model. At first glance, kdegi seemed less important for our experimentally assessed conditions. To further investigate the role of kdegi for indigoidine production kinetics, we systematically varied this parameter and compared simulation results. The synthesis of indigoidine was clearly affected by variation of kdegi (Fig. 7). The [Ind] trajectory collapsed more than 3-fold increase of the degradation rate. On the contrary, improved peptide stability could yield much more indigoidine. For very low degradation rates (i.e. kdegi < 1e-3), synthesis would not even saturate within the observed time frame.
Sensitivity analysis for optimized indigoidine production
To perceive the role of kdegi for indigoidine synthesis, we have varied the parameter over a broad range and observed qualitatively distinct model trajectories. For a more quantitative understanding of how indigoidine yield changes with altering parameters of the system, we conducted a sensitivity analysis[8]. Therefore, we first defined the variable IntInd, the integrated indigoidine reflecting production yield. The change of the indigoidine concentration with time is determined by the ODE \eqref{ind} and its integral with time can be depicted as the area under the [Ind] curve, i.e. the amount of indigoidine produced by the bacteria with time. Now we were interested in the change of IntInd with the change of the different parameters of our system. Mathematically, this is expressed as the so called sensitivity $\frac{\partial \mathrm{X}}{\partial \mathrm{p}}$, with X as our variable of interest (IntInd) and p as the parameter (beta, Bacmax, ksyn, kdim or kdegi, respectively). This partial derivative provides a measure of how much X changes with the change of p or how sensitive X is against small variations in p. Sensitivities can be scaled with the actual values of X and the respective p to fall in the range from -1 to +1. The higher the absolute value of the sensitivity, the more important is the parameter for the variable. A positive sensitivity means positive impact of the parameter on the variable whereas a sensitivity < 0 indicates reduction of X for subtle decrease in p. We utilised our mathematical model and computed the sensitivities of IntInd against the change of all five kinetic parameters of our system and scaled them accordingly. The analysis confirmed that the kdegi was the only parameter of the system that exhibited a negative impact on the indigoidine production (Tab. 1). Slight increase of all the other parameters also augmented additional yield.
Sensitivity analysis for native_sfp:| Parameter | $$\left.\mathrm{[Bac]}\right|_{t=0}$$ | kdegi | kdim | beta | ksyn | Bacmax |
|---|---|---|---|---|---|---|
| $$\frac{\partial\int\!\mathrm{Ind}\,\mathrm{d}t}{\partial p}$$ | 0.784 | -0.7024 | 0.5117 | 0.3338 | 0.2919 | 0.1036 |
The initial concentration of bacteria represented the most crucial parameter for indigoidine production, which was intuitive since the amount of produced indigoidine depends on the amount of bacteria in culture that synthesise the peptide. The parameter kdegi exhibited a high impact on IntInd but did not vary so much among the experimental conditions. One would assume that the half-life of the peptide is not affected by different T domains or PPTases. Possibly, the arrangement of modules influences peptide stability as inter-modular communication is a precondition for robust peptide assembly[9]. This links the peptide synthesis optimisation to our module shuffling project where modules of different hosts were combined for custom peptide synthesis. Indigoidine yield was also sensitive against small variations in beta and Bacmax. Culture conditions could be improved and metabolically more active strains could be chosen to further optimize performance in this respect. The parameters ksyn and kdim also determine peptide production.
The question is whether those rates could be altered by engineering the NRPS to advanced performance. To answer this question, we performed multiple linear regressions. We regressed the alignment score of the synthetic T domains of the indigoidine synthase with the rates from the respective best fit. The score was calculated by pairwise multiple sequence alignment of the amino acid sequences of the synthetic T domains with the native T domain of the indigoidine synthase as a template. The parameter values were obtained from parameter estimations of our mathematical model that has been calibrated with experimental data as described above. Linear regression of the alignment score on ksyn did only result in a coefficient of correlation of 0.48 while kdim correlated rather well (R2=0.97). This pinpointed the possibility to engineer T domains of NRPSs to optimize kinetic rates in silico for peptide production.
Conclusion and Outlook
We set up a mathematical model for bacterial indigoidine production to showcase the systematic scrutinization of NRP synthesis kinetics. In the course of model calibration with experimental data we could already falsify the hypothesis that indigoidine had an inhibitory effect on growth of the bacteria. The model with such inhibitory terms was not able to describe the experimental data. A reduced model was proven to be fully identifiable via a profile likelihood approach and simulations could reproduce experimental findings. The impact of the different rates on indigoidine production was investigated via sensitivity analysis. We can conclude that yield could be increased when peptide stability is improved and synthesis efficiency is advanced. Linear regression suggests that the dimerization rate of indigoidine could be engineered by editing the T domain sequence. With this, we have learnt valuable lessons on optimizing production of synthetic peptides in silico. Of course, dependency of yield on kinetic parameters does not necessarily scale linearly with peptide length but with these initial trials the way is paved for further analysis.
1. Brachmann, Alexander O, Kirchner, Ferdinand, Kegler, Carsten, Kinski, Sebastian C, Schmitt, Imke, Bode, Helge B: Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens., J. Biotechnol. 157(1), 96–9, January 2012
2. Klipp, Edda, Liebermeister, Wolfram: Mathematical modeling of intracellular signaling pathways., BMC Neurosci 7 Suppl 1, S10, 2006
3. Pruitt, Kenneth M., Kamau, David N.: Mathematical models of bacterial growth, inhibition and death under combined stress conditions, Journal of Industrial Microbiology 12(3-5), Springer, 221, 1993
4. Raue, Andreas, Schilling, Marcel, Bachmann, Julie, Matteson, Andrew, Schelke, Max, Kaschek, Daniel, Hug, Sabine, Kreutz, Clemens, Harms, Brian D, Theis, Fabian J, Klingmüller, Ursula, Timmer, Jens: Lessons learned from quantitative dynamical modeling in systems biology., PLoS ONE 8(9), e74335, 2013
5. Raue, A, Becker, V, Klingmüller, U, Timmer, J: Identifiability and observability analysis for experimental design in nonlinear dynamical models., Chaos 20(4), 045105, December 2010
6. Raue, A, Kreutz, C, Maiwald, T, Bachmann, J, Schilling, M, Klingmüller, U, Timmer, J: Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood., Bioinformatics 25(15), 1923–9, August 2009
7. Owen, Art B.: A Central Limit Theorem for Latin Hypercube Sampling, Journal of the Royal Statistical Society. Series B (Methodological) 54(2), Wiley for the Royal Statistical Society, 541–551, 1992
8. Zi, Zhike, Zheng, Yanan, Rundell, Ann E, Klipp, Edda: SBML-SAT: a systems biology markup language (SBML) based sensitivity analysis tool., BMC Bioinformatics 9, 342, 2008
9. Gokhale, R S, Tsuji, S Y, Cane, D E, Khosla, C: Dissecting and exploiting intermodular communication in polyketide synthases., Science 284(5413), 482–5, April 1999
 "
"