Team:Colombia Uniandes/NickoJournal
From 2013.igem.org
Camilog137 (Talk | contribs) |
Trafalmejo (Talk | contribs) |
||
| (63 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Http://2013.igem.org/Team:Colombia Uniandes/Bootstrap}} | {{Http://2013.igem.org/Team:Colombia Uniandes/Bootstrap}} | ||
| + | |||
| + | <div class="container"> | ||
| + | <div class="span11"> | ||
=='''Dear Journal! :)'''== | =='''Dear Journal! :)'''== | ||
| - | + | <p align="justify"> | |
Here we will place all the information about the work we have been doing during this time, it will be named by date. | Here we will place all the information about the work we have been doing during this time, it will be named by date. | ||
We hope you enjoy our work and thoughts as much as we do! | We hope you enjoy our work and thoughts as much as we do! | ||
| + | </p> | ||
[[File:La_foto.JPG|300px|thumb|center|Our Notebook]] | [[File:La_foto.JPG|300px|thumb|center|Our Notebook]] | ||
===='''18-Jun-2013'''==== | ===='''18-Jun-2013'''==== | ||
| - | + | <p align="justify"> | |
Hello! Hello! Hola! Hola! | Hello! Hello! Hola! Hola! | ||
| - | After we went over the project, we proceed to design our primers so we could place our order, taking into account that we are going to use Fusion PCR as our main protocol. Our primers take around 2 weeks to arrive… sooo meanwhilee all the team started making our ''E.coli'' babies electrocompetent, so we can work with them later on. | + | After we went over the project, we proceed to design our primers so we could place our order, taking into account that we are going to use Fusion PCR as our main protocol. If you want to check our primers, go to [https://2013.igem.org/Team:Colombia_Uniandes/How_to_parts How to: Parts]. Our primers take around 2 weeks to arrive… sooo meanwhilee all the team started making our ''E.coli'' babies electrocompetent, so we can work with them later on. |
| + | </p> | ||
===='''25-Jun-2013'''==== | ===='''25-Jun-2013'''==== | ||
| - | + | <p align="justify"> | |
So today we took our bacterial cells (Top 10) onto an LB medium (no antibiotics). We let it ON at 37 °C on the shaker. Also we prepared all the material we need for tomorrow: ddwater, 10 % glycerol, LB medium and microfuge tubes. | So today we took our bacterial cells (Top 10) onto an LB medium (no antibiotics). We let it ON at 37 °C on the shaker. Also we prepared all the material we need for tomorrow: ddwater, 10 % glycerol, LB medium and microfuge tubes. | ||
| - | + | </p> | |
===='''26-Jun-2013'''==== | ===='''26-Jun-2013'''==== | ||
| - | + | <p align="justify"> | |
Today we are going to start preparing our cells, we inoculated in the morning and placed all our material that needs to be cold in the fridge 4 °C(ddWater and glycerol), also the centifuge rotor needs to be chilled. | Today we are going to start preparing our cells, we inoculated in the morning and placed all our material that needs to be cold in the fridge 4 °C(ddWater and glycerol), also the centifuge rotor needs to be chilled. | ||
Then we just waited for the perfect OD and made our cells around 4:00pm. We made a lot! | Then we just waited for the perfect OD and made our cells around 4:00pm. We made a lot! | ||
| - | + | </p> | |
We placed them in the -80, ready to start our work!!! | We placed them in the -80, ready to start our work!!! | ||
| Line 28: | Line 33: | ||
So we made our plan(in theory)on big steps for the next weeks!: | So we made our plan(in theory)on big steps for the next weeks!: | ||
| - | '''1.''' '''DNA extraction''' from ''E.coli'' and '' | + | '''1.''' '''DNA extraction''' from ''E.coli'' and ''Cupriavidus metallidurans CH34'' |
'''2. PCR''' | '''2. PCR''' | ||
| Line 41: | Line 46: | ||
On each PCR add 30 steps more of making gels for confirmation, again and again and again. | On each PCR add 30 steps more of making gels for confirmation, again and again and again. | ||
| - | |||
| + | ===='''3-Jul-2013'''==== | ||
| + | <p align="justify"> | ||
Fortunately our team member Silvia Cañas already had a DNA extraction from ''E.coli'' so she donated it to us so we could start working! Thank you Silvia! :) | Fortunately our team member Silvia Cañas already had a DNA extraction from ''E.coli'' so she donated it to us so we could start working! Thank you Silvia! :) | ||
| - | We asked for '' | + | We asked for ''Cupriavidus metallidurans'' to our teacher Jenny Dussan at her lab here in the university. She is giving us the strain tomorrow morning! Thank you Jenny! |
| - | This done, did the extraction of '' | + | This done, did the extraction of ''C.metallidurans'' and store it in the -30 °C. |
| + | </p> | ||
===='''4-Jul-2013'''==== | ===='''4-Jul-2013'''==== | ||
| - | + | <p align="justify"> | |
Today we started with the 2-step PCR of PrcnR, RBS, ''rcnR'', stop (Fragment that will be called "R" from now on). | Today we started with the 2-step PCR of PrcnR, RBS, ''rcnR'', stop (Fragment that will be called "R" from now on). | ||
Results were negative :( | Results were negative :( | ||
| - | + | </p> | |
===='''5-Jul-2013'''==== | ===='''5-Jul-2013'''==== | ||
| - | + | <p align="justify"> | |
| - | Today we repeated "R" PCR and also did hoxN with the '' | + | Today we repeated "R" PCR and also did ''hoxN'' with the ''Cupriavidus metallidurans'' genome. The quantities used for the reaction of ''hoxN'' are shown in the table below. |
Results were negative :( as we could see in our sad, SAD gel. | Results were negative :( as we could see in our sad, SAD gel. | ||
| + | </p> | ||
| + | {| border="1" align="center" | ||
| + | |+'''PCR reagents and amounts for one 50 ul reaction''' | ||
| + | | style="text-align: center;" |Reagent | ||
| + | | style="text-align: center;" |Amount | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |''C.metallidurans'' DNA | ||
| + | | style="text-align: center;" |3,0 ul | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |Primer FW | ||
| + | | style="text-align: center;" |2,5 ul | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |Primer RV | ||
| + | | style="text-align: center;" |2,5 ul | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |DMSO | ||
| + | | style="text-align: center;" |2,0 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |Master Mix | ||
| + | | style="text-align: center;" |25 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |dH2O | ||
| + | | style="text-align: center;" |15 ul | ||
| + | |- | ||
| + | |} | ||
| + | |||
===='''9-Jul-2013'''==== | ===='''9-Jul-2013'''==== | ||
| + | <p align="justify"> | ||
| + | So, our PCRs are NOT working....We think our problem is the length of the primers and their constitution that could form secondary structures very VERY easily. | ||
| - | + | That's why we tried a new procotol of PCR for "R" with a temperature gradient, and we tried new buffers for ''hoxN''. Reagents and quantities are shown in the table below. | |
| + | </p> | ||
| + | {| border="1" align="center" | ||
| + | |+'''PCR reagents and amounts for one 50 ul reaction''' | ||
| + | | style="text-align: center;" |Reagent | ||
| + | | style="text-align: center;" |Amount | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |''Escherichia coli'' DNA | ||
| + | | style="text-align: center;" |2,0 ul | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |Primer FW | ||
| + | | style="text-align: center;" |2,5 ul | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |Primer RV | ||
| + | | style="text-align: center;" |2,5 ul | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |DMSO | ||
| + | | style="text-align: center;" |2,0 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |Master Mix | ||
| + | | style="text-align: center;" |25 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |dH2O | ||
| + | | style="text-align: center;" |16 ul | ||
| + | |- | ||
| + | |} | ||
| - | |||
We got a POSITIVE result on ''hoxN'', and POSITIVE with "R" at "low" temperatures of annealing. Here is our BEAUTIFULLL gel. | We got a POSITIVE result on ''hoxN'', and POSITIVE with "R" at "low" temperatures of annealing. Here is our BEAUTIFULLL gel. | ||
| Line 82: | Line 149: | ||
===='''15-Jul-2013'''==== | ===='''15-Jul-2013'''==== | ||
| - | We started ''hoxN'' PCR with the | + | We started ''hoxN'' PCR with the phusion primers, over the ''C. metallidurans'' genome, using 2-step PCR protocol. |
Results are negative. | Results are negative. | ||
| Line 92: | Line 159: | ||
Nothing worked OUT :( !!! | Nothing worked OUT :( !!! | ||
| - | We are going to try to do the PCR of hoxN with the | + | We are going to try to do the PCR of hoxN with the phusion primers over the fragment already amplified and see, as fas as PrcnA goes...we will try a temperature ramp with a 3-step PCR protocol. |
| - | + | ||
End for today :( | End for today :( | ||
| + | |||
| + | ===='''18-Jul-2013'''==== | ||
| + | |||
| + | Today we did ''E. coli'' chimiocompetent cells to mRFP transformation using TransforrmAid Bacterial Transformation Kit (Thermo Scientific). We did mRFP transformation from iGEM plate. | ||
| + | |||
| + | ===='''19-Jul-2013'''==== | ||
| + | We have transformants!!!! Beautiful, aren´t they? | ||
| + | [[File:IMG-20130909-00382.jpg|400px|thumb|center|Transformants]] | ||
| + | |||
| + | ===='''24-Jul-2013'''==== | ||
| + | <p align="justify"> | ||
| + | Today we are going to do pRcnA PCR. Besides iGEM kit, we used Phusion High Fidelity PCR kit and tested its two buffers: Phusion HF reaction Buffer and Phusion GC Reaction Buffer. We used a 2-step PCR protocol. Reagents and quantities of iGEM kit are the same we have been using until now, but reagents and quantities used for Phusion High Fidelity PCR kit are shown in the table below. | ||
| + | </p> | ||
| + | {| border="1" align="center" | ||
| + | |+'''PCR reagents and amounts for one 50 ul reaction''' | ||
| + | | style="text-align: center;" |Reagent | ||
| + | | style="text-align: center;" |Amount | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |''Escherichia coli'' DNA | ||
| + | | style="text-align: center;" |4,0 ul | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |Primer FW | ||
| + | | style="text-align: center;" |2,5 ul | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |Primer RV | ||
| + | | style="text-align: center;" |2,5 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |Buffer | ||
| + | | style="text-align: center;" |10 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |dNTPs | ||
| + | | style="text-align: center;" |1,0 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |DMSO | ||
| + | | style="text-align: center;" |1,5 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |Pol | ||
| + | | style="text-align: center;" |0,5 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |dH2O | ||
| + | | style="text-align: center;" |28,3 ul | ||
| + | |- | ||
| + | |} | ||
| + | ===='''25-Jul-2013'''==== | ||
| + | We did ''hoxN'' PCR by using the same kits than yesterday and 2-steps protocol. However, results are negative :'(. | ||
| + | |||
| + | ===='''26-Jul-2013'''==== | ||
| + | <p align="justify"> | ||
| + | Since we have been having troubles with PCRs, we decided to do genome extraction of our bacteria again. For this purpose we used Easy DNA Isolation Kit, Invitrogen. We followed manufacturer's instructions. We confirmed the extraction by gel electrophoresis. Genomes of ''C. metallidurans'' and new ''E.coli'' Extracted :D | ||
| + | |||
| + | [[File:GelGenoma.jpg|400px|thumb|center|GenomeNewExtractionWithOUTRnase]] | ||
| + | |||
| + | </p> | ||
| + | |||
| + | *Remember you can review protocols in the section named [https://2013.igem.org/Team:Colombia_Uniandes/Protocols Protocols] | ||
| + | |||
| + | ===='''29-Jul-2013'''==== | ||
| + | Today we did PCR of ''hoxN'' from the new ''C. metallidurans'' genome exracted and guess what? We got ''hoxN'' a beatiful band around 600 pb :D :D. Blue arrow shows HoxN and yellow arrow shows rcnR...FAIL | ||
| + | |||
| + | [[File:GelHoxN.jpg|400px|thumb|center|GelHoxN]] | ||
| + | |||
| + | ===='''30-Jul-2013'''==== | ||
| + | We did PCR of ''rcnR'' using the ''E. coli'' genome extracted last Friday and Phusion High Fidelity PCR kit. On the gel we got a diffuse band around 400 pb, so we could not conclude anything. | ||
| + | |||
| + | ===='''31-Jul-2013'''==== | ||
| + | <p align="justify"> | ||
| + | We did PCR of ''pRcnA'' and ''rcnR'' again using a annealing temperature gradient between 45°C and 60° and using the two buffers: Phusion HF reaction Buffer and Phusion GC Reaction Buffer. On the gel we saw a band that corresponded to ''pRcnA'' when the annealing temperature had been around 53°C and GC buffer had been used. On the other hand, in ''rcnR'' reactions we got a band when buffer HF had been used and when annealing temperature was around 58°C. | ||
| + | So...u CAN BARELY see them BUT THEY ARE THERE! | ||
| + | |||
| + | [[File:PrcnA.jpg|400px|thumb|center|Prcna,rcnR]] | ||
| + | </p> | ||
| + | |||
| + | ===='''01-Aug-2013'''==== | ||
| + | <p align="justify"> | ||
| + | Today we did ''hoxN''-''rfp'' phusion. We used Phusion High Fidelity PCR kit, Phusion HF reaction Buffer and the amounts of other reagents described below. We got our first phusion!!! What a great day :) | ||
| + | </p> | ||
| + | {| border="1" align="center" | ||
| + | |+'''Phusion PCR''' | ||
| + | | style="text-align: center;" |Reagent | ||
| + | | style="text-align: center;" |Amount | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |''rfp'' DNA | ||
| + | | style="text-align: center;" |2,0 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |''hoxN'' DNA | ||
| + | | style="text-align: center;" |2,0 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |Primer FW | ||
| + | | style="text-align: center;" |1,0 ul | ||
| + | |||
| + | |- | ||
| + | | style="text-align: center;" |Primer RV | ||
| + | | style="text-align: center;" |1,0 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |Buffer | ||
| + | | style="text-align: center;" |4 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |dNTPs | ||
| + | | style="text-align: center;" |0,4 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |DMSO | ||
| + | | style="text-align: center;" |1,0 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |Pol | ||
| + | | style="text-align: center;" |0,2 ul | ||
| + | |- | ||
| + | | style="text-align: center;" |dH2O | ||
| + | | style="text-align: center;" |8,4 ul | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | [[File:hoxNRFP.jpg|400px|thumb|center|hoxNRFP fusion!]] | ||
| + | |||
| + | ===='''02-Aug-2013'''==== | ||
| + | Another try of PrcnR-rcnR and then again...nothing. | ||
| + | |||
| + | [[File:rcnR.jpg|400px|thumb|center|rcnR]] | ||
| + | |||
| + | ===='''05-Aug-2013'''==== | ||
| + | |||
| + | Since we obtained PrcnA and HoxN, we made a Prcna-HoxN fusion and we also amplified hoxN-RFP fusion. All was obtained and we can see that in our beautifull gel :)Go guys!! The fisrt column next to the DNA ladder is HoxN-RFP fusion on the first pink arrow, on the second pink arrow there is our fusion Prcna-HoxN. | ||
| + | |||
| + | [[File:Prcna_hoxN.jpg|400px|thumb|center|Prcna-HoxN and HoxN-RFP]] | ||
| + | |||
| + | |||
| + | ===='''07-Aug-2013'''==== | ||
| + | |||
| + | Today we decided to digest our parts with EcoRI in order to insert our parts in the backbone. Thats why first we did the digestion with EcoRI on the Backbone and also the same process with the parts. We digested for 2 hours and then desactivated the ennzymes at 80°C for 20 min. For protocol see: https://www.neb.com/products/r0101-ecori#tabselect2 | ||
| + | |||
| + | We then used antarctic phosphatase for the backbone for 1h. For protocol see: https://www.neb.com/protocols/1/01/01/vector-dephosphorylation-protocol | ||
| + | |||
| + | Finally we let the ligation all night at room temperature. | ||
| + | |||
| + | ===='''09-Aug-2013'''==== | ||
| + | |||
| + | Today we desactivated the ligase at 80°C for 15 min and started THE TRANSFORMATION! | ||
| + | |||
| + | We transformated our cells and let them in 37°C. Lets see tomorrow :) | ||
| + | |||
| + | ===='''10-Aug-2013'''==== | ||
| + | They were NO colonies At ALL! not even halfffffff, not even a tiny oneee!! | ||
| + | |||
| + | We are repeating process on monday. | ||
| + | |||
| + | ===='''13-Aug-2013'''==== | ||
| + | Today we did electrocompetent cells with E.coli DH10B and we tranformed and well...hopefully we will see something tomorrow! | ||
| + | |||
| + | Hopes up! | ||
| + | |||
| + | ===='''14-Aug-2013'''==== | ||
| + | |||
| + | We have some colonies! but only on HoxN-RFP fusion! We are going to leave those on ON and we are transforming Prcna-HoxN again. | ||
| + | |||
| + | ===='''15-Aug-2013'''==== | ||
| + | |||
| + | Transformation not succesfull, do it one more time! | ||
| + | |||
| + | ===='''16-Aug-2013'''==== | ||
| + | |||
| + | Transformation not succesfull, do it one more time! | ||
| + | |||
| + | ===='''19-Aug-2013'''==== | ||
| + | |||
| + | Transformation not succesfull, do it one more time! | ||
| + | |||
| + | |||
| + | ===='''20-Aug-2013'''==== | ||
| + | |||
| + | Tranformation succesfull!!!! :) Growing on ON! | ||
| + | |||
| + | ===='''21-Aug-2013'''==== | ||
| + | |||
| + | Miniprep DAY! :) | ||
| + | |||
| + | Parts Ready! | ||
| + | |||
| + | ===='''23-Aug-2013'''==== | ||
| + | |||
| + | We are sending Prcna-HoxN and HoxN-RFP :) | ||
| + | We are still trying to get Prcnr-rcnR so lets go! | ||
| + | |||
| + | ===='''24-Aug-2013'''==== | ||
| + | |||
| + | We tried PCR Prcnr-rcnR and IS NOT THEREEEEEEEEEEEE! | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
Latest revision as of 15:27, 26 September 2013
Dear Journal! :)
Here we will place all the information about the work we have been doing during this time, it will be named by date.
We hope you enjoy our work and thoughts as much as we do!
18-Jun-2013
Hello! Hello! Hola! Hola!
After we went over the project, we proceed to design our primers so we could place our order, taking into account that we are going to use Fusion PCR as our main protocol. If you want to check our primers, go to How to: Parts. Our primers take around 2 weeks to arrive… sooo meanwhilee all the team started making our E.coli babies electrocompetent, so we can work with them later on.
25-Jun-2013
So today we took our bacterial cells (Top 10) onto an LB medium (no antibiotics). We let it ON at 37 °C on the shaker. Also we prepared all the material we need for tomorrow: ddwater, 10 % glycerol, LB medium and microfuge tubes.
26-Jun-2013
Today we are going to start preparing our cells, we inoculated in the morning and placed all our material that needs to be cold in the fridge 4 °C(ddWater and glycerol), also the centifuge rotor needs to be chilled.
Then we just waited for the perfect OD and made our cells around 4:00pm. We made a lot!
We placed them in the -80, ready to start our work!!!
2-Jul-2013
Our primers are FINALLY here and we are anxious to start working!!!
So we made our plan(in theory)on big steps for the next weeks!:
1. DNA extraction from E.coli and Cupriavidus metallidurans CH34
2. PCR
2.1 PrcnR, RBS, rcnR, stop (Amplified as one piece)
2.2 PrcnA,RBS
2.3 hoxN, stop
3. Fusion PCR
3.1 PrcnA/hoxN
3.2 PrcnR,RBS,rcnR/PrcnA/hoxN
On each PCR add 30 steps more of making gels for confirmation, again and again and again.
3-Jul-2013
Fortunately our team member Silvia Cañas already had a DNA extraction from E.coli so she donated it to us so we could start working! Thank you Silvia! :)
We asked for Cupriavidus metallidurans to our teacher Jenny Dussan at her lab here in the university. She is giving us the strain tomorrow morning! Thank you Jenny!
This done, did the extraction of C.metallidurans and store it in the -30 °C.
4-Jul-2013
Today we started with the 2-step PCR of PrcnR, RBS, rcnR, stop (Fragment that will be called "R" from now on).
Results were negative :(
5-Jul-2013
Today we repeated "R" PCR and also did hoxN with the Cupriavidus metallidurans genome. The quantities used for the reaction of hoxN are shown in the table below.
Results were negative :( as we could see in our sad, SAD gel.
PCR reagents and amounts for one 50 ul reaction
Reagent
Amount
C.metallidurans DNA
3,0 ul
Primer FW
2,5 ul
Primer RV
2,5 ul
DMSO
2,0 ul
Master Mix
25 ul
dH2O
15 ul
9-Jul-2013
So, our PCRs are NOT working....We think our problem is the length of the primers and their constitution that could form secondary structures very VERY easily.
That's why we tried a new procotol of PCR for "R" with a temperature gradient, and we tried new buffers for hoxN. Reagents and quantities are shown in the table below.
PCR reagents and amounts for one 50 ul reaction
Reagent
Amount
Escherichia coli DNA
2,0 ul
Primer FW
2,5 ul
Primer RV
2,5 ul
DMSO
2,0 ul
Master Mix
25 ul
dH2O
16 ul
We got a POSITIVE result on hoxN, and POSITIVE with "R" at "low" temperatures of annealing. Here is our BEAUTIFULLL gel.
On 5,7,8,9 we can see "R" fragment of 359bp.
On 12 we can see hoxN fragment of 837bp.
11-Jul-2013
Today we did the PrcnA PCR and it didn't work out.
15-Jul-2013
We started hoxN PCR with the phusion primers, over the C. metallidurans genome, using 2-step PCR protocol.
Results are negative.
17-Jul-2013
We repeated hoxN PCR with the fusion primers over the genome and PrcnA PCR with E.coli genome.
Nothing worked OUT :( !!!
We are going to try to do the PCR of hoxN with the phusion primers over the fragment already amplified and see, as fas as PrcnA goes...we will try a temperature ramp with a 3-step PCR protocol.
End for today :(
18-Jul-2013
Today we did E. coli chimiocompetent cells to mRFP transformation using TransforrmAid Bacterial Transformation Kit (Thermo Scientific). We did mRFP transformation from iGEM plate.
19-Jul-2013
We have transformants!!!! Beautiful, aren´t they?
24-Jul-2013
Today we are going to do pRcnA PCR. Besides iGEM kit, we used Phusion High Fidelity PCR kit and tested its two buffers: Phusion HF reaction Buffer and Phusion GC Reaction Buffer. We used a 2-step PCR protocol. Reagents and quantities of iGEM kit are the same we have been using until now, but reagents and quantities used for Phusion High Fidelity PCR kit are shown in the table below.
PCR reagents and amounts for one 50 ul reaction
Reagent
Amount
Escherichia coli DNA
4,0 ul
Primer FW
2,5 ul
Primer RV
2,5 ul
Buffer
10 ul
dNTPs
1,0 ul
DMSO
1,5 ul
Pol
0,5 ul
dH2O
28,3 ul
25-Jul-2013
We did hoxN PCR by using the same kits than yesterday and 2-steps protocol. However, results are negative :'(.
26-Jul-2013
Since we have been having troubles with PCRs, we decided to do genome extraction of our bacteria again. For this purpose we used Easy DNA Isolation Kit, Invitrogen. We followed manufacturer's instructions. We confirmed the extraction by gel electrophoresis. Genomes of C. metallidurans and new E.coli Extracted :D
- Remember you can review protocols in the section named Protocols
29-Jul-2013
Today we did PCR of hoxN from the new C. metallidurans genome exracted and guess what? We got hoxN a beatiful band around 600 pb :D :D. Blue arrow shows HoxN and yellow arrow shows rcnR...FAIL
30-Jul-2013
We did PCR of rcnR using the E. coli genome extracted last Friday and Phusion High Fidelity PCR kit. On the gel we got a diffuse band around 400 pb, so we could not conclude anything.
31-Jul-2013
We did PCR of pRcnA and rcnR again using a annealing temperature gradient between 45°C and 60° and using the two buffers: Phusion HF reaction Buffer and Phusion GC Reaction Buffer. On the gel we saw a band that corresponded to pRcnA when the annealing temperature had been around 53°C and GC buffer had been used. On the other hand, in rcnR reactions we got a band when buffer HF had been used and when annealing temperature was around 58°C.
So...u CAN BARELY see them BUT THEY ARE THERE!
01-Aug-2013
Today we did hoxN-rfp phusion. We used Phusion High Fidelity PCR kit, Phusion HF reaction Buffer and the amounts of other reagents described below. We got our first phusion!!! What a great day :)
Phusion PCR
Reagent
Amount
rfp DNA
2,0 ul
hoxN DNA
2,0 ul
Primer FW
1,0 ul
Primer RV
1,0 ul
Buffer
4 ul
dNTPs
0,4 ul
DMSO
1,0 ul
Pol
0,2 ul
dH2O
8,4 ul
02-Aug-2013
Another try of PrcnR-rcnR and then again...nothing.
05-Aug-2013
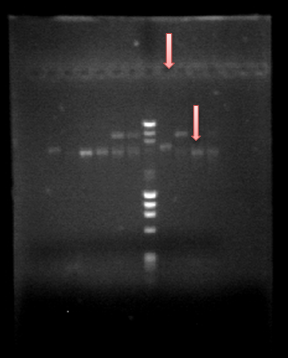
Since we obtained PrcnA and HoxN, we made a Prcna-HoxN fusion and we also amplified hoxN-RFP fusion. All was obtained and we can see that in our beautifull gel :)Go guys!! The fisrt column next to the DNA ladder is HoxN-RFP fusion on the first pink arrow, on the second pink arrow there is our fusion Prcna-HoxN.
07-Aug-2013
Today we decided to digest our parts with EcoRI in order to insert our parts in the backbone. Thats why first we did the digestion with EcoRI on the Backbone and also the same process with the parts. We digested for 2 hours and then desactivated the ennzymes at 80°C for 20 min. For protocol see: https://www.neb.com/products/r0101-ecori#tabselect2
We then used antarctic phosphatase for the backbone for 1h. For protocol see: https://www.neb.com/protocols/1/01/01/vector-dephosphorylation-protocol
Finally we let the ligation all night at room temperature.
09-Aug-2013
Today we desactivated the ligase at 80°C for 15 min and started THE TRANSFORMATION!
We transformated our cells and let them in 37°C. Lets see tomorrow :)
10-Aug-2013
They were NO colonies At ALL! not even halfffffff, not even a tiny oneee!!
We are repeating process on monday.
13-Aug-2013
Today we did electrocompetent cells with E.coli DH10B and we tranformed and well...hopefully we will see something tomorrow!
Hopes up!
14-Aug-2013
We have some colonies! but only on HoxN-RFP fusion! We are going to leave those on ON and we are transforming Prcna-HoxN again.
15-Aug-2013
Transformation not succesfull, do it one more time!
16-Aug-2013
Transformation not succesfull, do it one more time!
19-Aug-2013
Transformation not succesfull, do it one more time!
20-Aug-2013
Tranformation succesfull!!!! :) Growing on ON!
21-Aug-2013
Miniprep DAY! :)
Parts Ready!
23-Aug-2013
We are sending Prcna-HoxN and HoxN-RFP :)
We are still trying to get Prcnr-rcnR so lets go!
24-Aug-2013
We tried PCR Prcnr-rcnR and IS NOT THEREEEEEEEEEEEE!
Dear Journal! :)
Here we will place all the information about the work we have been doing during this time, it will be named by date. We hope you enjoy our work and thoughts as much as we do!
18-Jun-2013
Hello! Hello! Hola! Hola! After we went over the project, we proceed to design our primers so we could place our order, taking into account that we are going to use Fusion PCR as our main protocol. If you want to check our primers, go to How to: Parts. Our primers take around 2 weeks to arrive… sooo meanwhilee all the team started making our E.coli babies electrocompetent, so we can work with them later on.
25-Jun-2013
So today we took our bacterial cells (Top 10) onto an LB medium (no antibiotics). We let it ON at 37 °C on the shaker. Also we prepared all the material we need for tomorrow: ddwater, 10 % glycerol, LB medium and microfuge tubes.
26-Jun-2013
Today we are going to start preparing our cells, we inoculated in the morning and placed all our material that needs to be cold in the fridge 4 °C(ddWater and glycerol), also the centifuge rotor needs to be chilled. Then we just waited for the perfect OD and made our cells around 4:00pm. We made a lot!
We placed them in the -80, ready to start our work!!!
2-Jul-2013
Our primers are FINALLY here and we are anxious to start working!!! So we made our plan(in theory)on big steps for the next weeks!:
1. DNA extraction from E.coli and Cupriavidus metallidurans CH34
2. PCR
2.1 PrcnR, RBS, rcnR, stop (Amplified as one piece) 2.2 PrcnA,RBS 2.3 hoxN, stop
3. Fusion PCR
3.1 PrcnA/hoxN 3.2 PrcnR,RBS,rcnR/PrcnA/hoxN
On each PCR add 30 steps more of making gels for confirmation, again and again and again.
3-Jul-2013
Fortunately our team member Silvia Cañas already had a DNA extraction from E.coli so she donated it to us so we could start working! Thank you Silvia! :) We asked for Cupriavidus metallidurans to our teacher Jenny Dussan at her lab here in the university. She is giving us the strain tomorrow morning! Thank you Jenny! This done, did the extraction of C.metallidurans and store it in the -30 °C.
4-Jul-2013
Today we started with the 2-step PCR of PrcnR, RBS, rcnR, stop (Fragment that will be called "R" from now on). Results were negative :(
5-Jul-2013
Today we repeated "R" PCR and also did hoxN with the Cupriavidus metallidurans genome. The quantities used for the reaction of hoxN are shown in the table below. Results were negative :( as we could see in our sad, SAD gel.
| Reagent | Amount |
| C.metallidurans DNA | 3,0 ul |
| Primer FW | 2,5 ul |
| Primer RV | 2,5 ul |
| DMSO | 2,0 ul |
| Master Mix | 25 ul |
| dH2O | 15 ul |
9-Jul-2013
So, our PCRs are NOT working....We think our problem is the length of the primers and their constitution that could form secondary structures very VERY easily. That's why we tried a new procotol of PCR for "R" with a temperature gradient, and we tried new buffers for hoxN. Reagents and quantities are shown in the table below.
| Reagent | Amount |
| Escherichia coli DNA | 2,0 ul |
| Primer FW | 2,5 ul |
| Primer RV | 2,5 ul |
| DMSO | 2,0 ul |
| Master Mix | 25 ul |
| dH2O | 16 ul |
We got a POSITIVE result on hoxN, and POSITIVE with "R" at "low" temperatures of annealing. Here is our BEAUTIFULLL gel.
On 5,7,8,9 we can see "R" fragment of 359bp. On 12 we can see hoxN fragment of 837bp.
11-Jul-2013
Today we did the PrcnA PCR and it didn't work out.
15-Jul-2013
We started hoxN PCR with the phusion primers, over the C. metallidurans genome, using 2-step PCR protocol.
Results are negative.
17-Jul-2013
We repeated hoxN PCR with the fusion primers over the genome and PrcnA PCR with E.coli genome.
Nothing worked OUT :( !!!
We are going to try to do the PCR of hoxN with the phusion primers over the fragment already amplified and see, as fas as PrcnA goes...we will try a temperature ramp with a 3-step PCR protocol. End for today :(
18-Jul-2013
Today we did E. coli chimiocompetent cells to mRFP transformation using TransforrmAid Bacterial Transformation Kit (Thermo Scientific). We did mRFP transformation from iGEM plate.
19-Jul-2013
We have transformants!!!! Beautiful, aren´t they?
24-Jul-2013
Today we are going to do pRcnA PCR. Besides iGEM kit, we used Phusion High Fidelity PCR kit and tested its two buffers: Phusion HF reaction Buffer and Phusion GC Reaction Buffer. We used a 2-step PCR protocol. Reagents and quantities of iGEM kit are the same we have been using until now, but reagents and quantities used for Phusion High Fidelity PCR kit are shown in the table below.
| Reagent | Amount |
| Escherichia coli DNA | 4,0 ul |
| Primer FW | 2,5 ul |
| Primer RV | 2,5 ul |
| Buffer | 10 ul |
| dNTPs | 1,0 ul |
| DMSO | 1,5 ul |
| Pol | 0,5 ul |
| dH2O | 28,3 ul |
25-Jul-2013
We did hoxN PCR by using the same kits than yesterday and 2-steps protocol. However, results are negative :'(.
26-Jul-2013
Since we have been having troubles with PCRs, we decided to do genome extraction of our bacteria again. For this purpose we used Easy DNA Isolation Kit, Invitrogen. We followed manufacturer's instructions. We confirmed the extraction by gel electrophoresis. Genomes of C. metallidurans and new E.coli Extracted :D
- Remember you can review protocols in the section named Protocols
29-Jul-2013
Today we did PCR of hoxN from the new C. metallidurans genome exracted and guess what? We got hoxN a beatiful band around 600 pb :D :D. Blue arrow shows HoxN and yellow arrow shows rcnR...FAIL
30-Jul-2013
We did PCR of rcnR using the E. coli genome extracted last Friday and Phusion High Fidelity PCR kit. On the gel we got a diffuse band around 400 pb, so we could not conclude anything.
31-Jul-2013
We did PCR of pRcnA and rcnR again using a annealing temperature gradient between 45°C and 60° and using the two buffers: Phusion HF reaction Buffer and Phusion GC Reaction Buffer. On the gel we saw a band that corresponded to pRcnA when the annealing temperature had been around 53°C and GC buffer had been used. On the other hand, in rcnR reactions we got a band when buffer HF had been used and when annealing temperature was around 58°C. So...u CAN BARELY see them BUT THEY ARE THERE!
01-Aug-2013
Today we did hoxN-rfp phusion. We used Phusion High Fidelity PCR kit, Phusion HF reaction Buffer and the amounts of other reagents described below. We got our first phusion!!! What a great day :)
| Reagent | Amount |
| rfp DNA | 2,0 ul |
| hoxN DNA | 2,0 ul |
| Primer FW | 1,0 ul |
| Primer RV | 1,0 ul |
| Buffer | 4 ul |
| dNTPs | 0,4 ul |
| DMSO | 1,0 ul |
| Pol | 0,2 ul |
| dH2O | 8,4 ul |
02-Aug-2013
Another try of PrcnR-rcnR and then again...nothing.
05-Aug-2013
Since we obtained PrcnA and HoxN, we made a Prcna-HoxN fusion and we also amplified hoxN-RFP fusion. All was obtained and we can see that in our beautifull gel :)Go guys!! The fisrt column next to the DNA ladder is HoxN-RFP fusion on the first pink arrow, on the second pink arrow there is our fusion Prcna-HoxN.
07-Aug-2013
Today we decided to digest our parts with EcoRI in order to insert our parts in the backbone. Thats why first we did the digestion with EcoRI on the Backbone and also the same process with the parts. We digested for 2 hours and then desactivated the ennzymes at 80°C for 20 min. For protocol see: https://www.neb.com/products/r0101-ecori#tabselect2
We then used antarctic phosphatase for the backbone for 1h. For protocol see: https://www.neb.com/protocols/1/01/01/vector-dephosphorylation-protocol
Finally we let the ligation all night at room temperature.
09-Aug-2013
Today we desactivated the ligase at 80°C for 15 min and started THE TRANSFORMATION!
We transformated our cells and let them in 37°C. Lets see tomorrow :)
10-Aug-2013
They were NO colonies At ALL! not even halfffffff, not even a tiny oneee!!
We are repeating process on monday.
13-Aug-2013
Today we did electrocompetent cells with E.coli DH10B and we tranformed and well...hopefully we will see something tomorrow!
Hopes up!
14-Aug-2013
We have some colonies! but only on HoxN-RFP fusion! We are going to leave those on ON and we are transforming Prcna-HoxN again.
15-Aug-2013
Transformation not succesfull, do it one more time!
16-Aug-2013
Transformation not succesfull, do it one more time!
19-Aug-2013
Transformation not succesfull, do it one more time!
20-Aug-2013
Tranformation succesfull!!!! :) Growing on ON!
21-Aug-2013
Miniprep DAY! :)
Parts Ready!
23-Aug-2013
We are sending Prcna-HoxN and HoxN-RFP :) We are still trying to get Prcnr-rcnR so lets go!
24-Aug-2013
We tried PCR Prcnr-rcnR and IS NOT THEREEEEEEEEEEEE!
 "
"