Team:DTU-Denmark/Notebook/23 July 2013
From 2013.igem.org
(→208) |
|||
| (11 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:DTU-Denmark/Templates/StartPage| | + | {{:Team:DTU-Denmark/Templates/StartPage|23 July 2013}} |
| - | =208= | + | Navigate to the [[Team:DTU-Denmark/Notebook/22_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/24_July_2013|Next]] Entry |
| + | =Lab 208= | ||
<hr/> | <hr/> | ||
==Main purpose== | ==Main purpose== | ||
| Line 7: | Line 8: | ||
==Who was in the lab== | ==Who was in the lab== | ||
| + | <hr/> | ||
Gosia, Kristian, Henrike | Gosia, Kristian, Henrike | ||
| - | |||
==Procedure== | ==Procedure== | ||
| + | <hr/> | ||
| + | ===Gel casting and loading=== | ||
| + | |||
| + | 1% agarose gels were prepared with 5% Ethidium bromide. | ||
==Results== | ==Results== | ||
<hr/> | <hr/> | ||
===Verification gel=== | ===Verification gel=== | ||
| - | |||
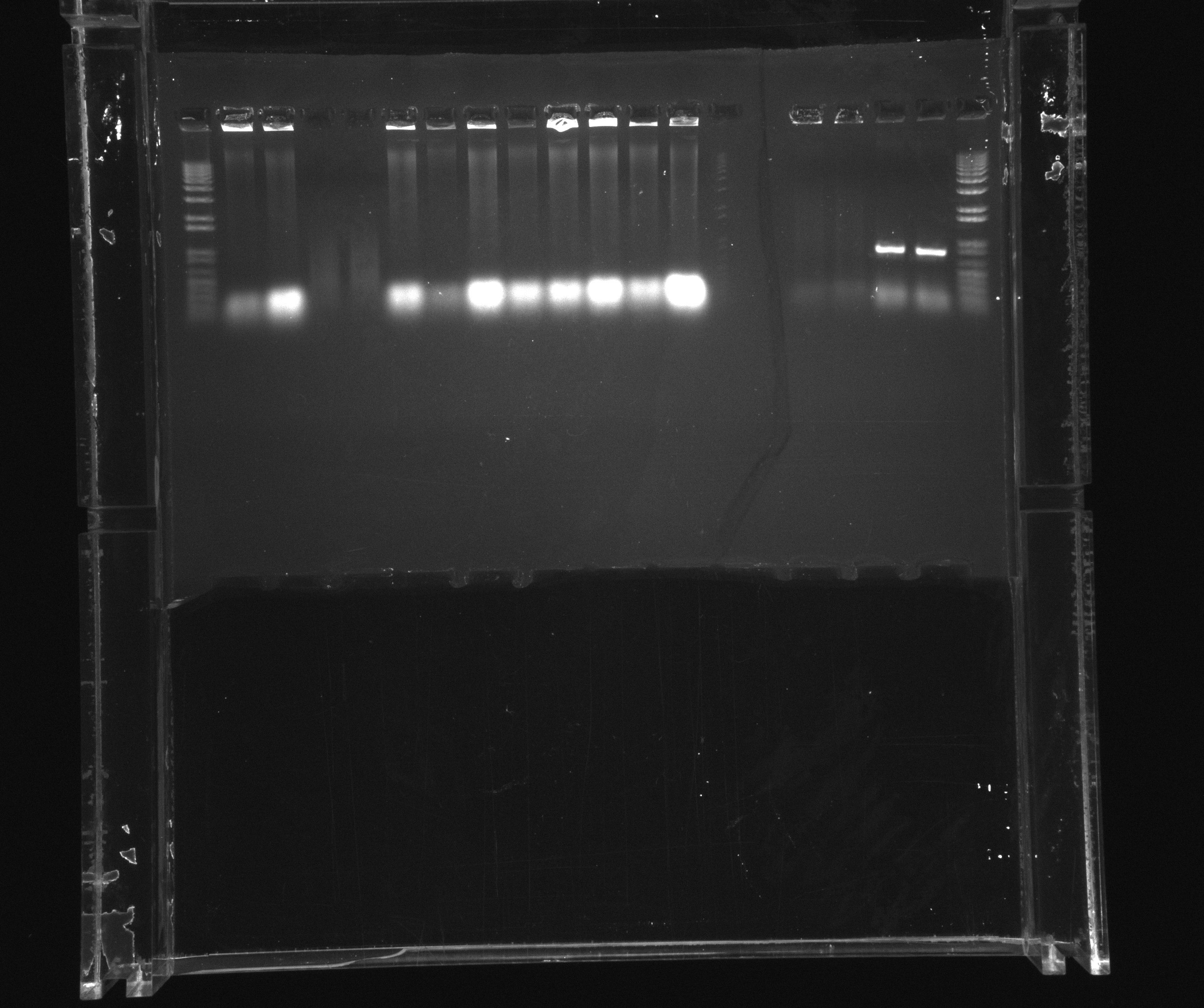
1% gel to verify our earlier PCR purifications | 1% gel to verify our earlier PCR purifications | ||
| Line 22: | Line 26: | ||
*1: broad band ladder | *1: broad band ladder | ||
| - | *2: HAO extracted from N. | + | *2: HAO extracted from ''N. europaea'' with flanking primers (A4) |
| - | *3: AMO extracted from N. | + | *3: AMO extracted from ''N. europaea'' with flanking primers (A6) |
| - | *4: cycAX extracted from N. | + | *4: cycAX extracted from ''N. europaea'' with flanking primers (A5) |
| - | *5: Nir extracted from P. aeruginosa with flanking primers (A7) | + | *5: Nir extracted from ''P. aeruginosa'' with flanking primers (A7) |
| - | *6: Nir extracted from P. aeruginosa with flanking primers (A8) | + | *6: Nir extracted from ''P. aeruginosa'' with flanking primers (A8) |
| - | *7: Nir extracted from P. aeruginosa with flanking primers (B8) | + | *7: Nir extracted from ''P. aeruginosa'' with flanking primers (B8) |
| - | *8: Nir extracted from P. aeruginosa with flanking primers (B9) | + | *8: Nir extracted from ''P. aeruginosa'' with flanking primers (B9) |
*9: pZA21 USER fragment, DpnI treated (F1) | *9: pZA21 USER fragment, DpnI treated (F1) | ||
*10: cycAX USER fragment (G1) | *10: cycAX USER fragment (G1) | ||
| Line 67: | Line 71: | ||
==Conclusions== | ==Conclusions== | ||
| + | <hr/> | ||
We USER-procucts seems to be fine and we then speculate that the reason why we only see cycAX as being successful in the transformation is due the destroying effect of too many membrane proteins. We will try to solve this by putting HAO and AMO under an inducible promoter. | We USER-procucts seems to be fine and we then speculate that the reason why we only see cycAX as being successful in the transformation is due the destroying effect of too many membrane proteins. We will try to solve this by putting HAO and AMO under an inducible promoter. | ||
The pZA21::RFP transformation seems to be successful on colony-PCR and this is also visual from the red colonies. | The pZA21::RFP transformation seems to be successful on colony-PCR and this is also visual from the red colonies. | ||
Latest revision as of 22:03, 29 September 2013
23 July 2013
Contents |
Lab 208
Main purpose
Find out what is wrong with Nir-extraction and why we make a successful USER-reaction with any USER-products from HAO and AMO.
Who was in the lab
Gosia, Kristian, Henrike
Procedure
Gel casting and loading
1% agarose gels were prepared with 5% Ethidium bromide.
Results
Verification gel
1% gel to verify our earlier PCR purifications
wells (in bracets position of the purification in the gene box):
- 1: broad band ladder
- 2: HAO extracted from N. europaea with flanking primers (A4)
- 3: AMO extracted from N. europaea with flanking primers (A6)
- 4: cycAX extracted from N. europaea with flanking primers (A5)
- 5: Nir extracted from P. aeruginosa with flanking primers (A7)
- 6: Nir extracted from P. aeruginosa with flanking primers (A8)
- 7: Nir extracted from P. aeruginosa with flanking primers (B8)
- 8: Nir extracted from P. aeruginosa with flanking primers (B9)
- 9: pZA21 USER fragment, DpnI treated (F1)
- 10: cycAX USER fragment (G1)
- 11: AMO USER fragment (G2)
- 12: HAO USER fragment (G3)
- 13: Nir USER fragment (G4)
- 14: AMO USER fragment (G5)
- 15: 1kb plus ladder
colony PCR gel
1% gel to analyse yesterday's colony PCR
wells:
- 1: 1 kb ladder
- 2: HAO colony 1
- 3: HAO colony 2
- 4: RFP in pZA21 without promoter
- 5: RFP in pZA21 without promoter
- 6: AMO colony 3 (count starts from 3)
- 7: AMO colony 4
- 8: AMO colony 5
- 9: AMO colony 6
- 10: AMO colony 7
- 11: AMO colony 8
- 12: AMO colony 9
- 13: AMO colony 10
- break in the gel
- 14: cycAX colony 1
- 15: cycAX colony 2
- 14: RFP colony 1
- 14: RFP colony 2
Conclusions
We USER-procucts seems to be fine and we then speculate that the reason why we only see cycAX as being successful in the transformation is due the destroying effect of too many membrane proteins. We will try to solve this by putting HAO and AMO under an inducible promoter. The pZA21::RFP transformation seems to be successful on colony-PCR and this is also visual from the red colonies.
Navigate to the Previous or the Next Entry
 "
"