Team:DTU-Denmark/Notebook/29 July 2013
From 2013.igem.org
(→Main purpose) |
|||
| (27 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:DTU-Denmark/Templates/StartPage|29 July 2013}} | {{:Team:DTU-Denmark/Templates/StartPage|29 July 2013}} | ||
| - | + | Navigate to the [[Team:DTU-Denmark/Notebook/28_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/30_July_2013|Next]] Entry | |
| - | = | + | =Lab 208= |
<hr/> | <hr/> | ||
==Main purpose== | ==Main purpose== | ||
<hr/> | <hr/> | ||
| - | *Colony PCR from ''Pseudomonas aeruginosa'' ( | + | *Colony PCR from ''Pseudomonas aeruginosa'' (PAO1) to isolate Nir. |
| - | *Colony PCR to check the presence of AMO and HAO in | + | *Colony PCR to check the presence of AMO and HAO in transformants ''E.coli''. |
| + | Gel on yesterdays PCR | ||
| + | Isolation of genomic DNA from PAO1 | ||
==Who was in the lab== | ==Who was in the lab== | ||
| Line 14: | Line 16: | ||
==Procedure== | ==Procedure== | ||
<hr/> | <hr/> | ||
| + | ===Colony PCR=== | ||
| + | [[Team:DTU-Denmark/Methods/Colony_PCR|Colony PCR]] was performed according to standard method in order to check if transformed cells which grew on medium contain desired insert in pZA21 plasmid. | ||
| + | The colonies were chosen from agar plate to be checked. | ||
| + | Names and numbers of colonies as well as plates names and primers are: | ||
| + | |||
| + | * HAO 1-6 in pZA21 USER, primers 28a, 28b | ||
| + | * AMO 1-9 in pZA21 USER, primers 29a, 29b | ||
| + | |||
| + | |||
| + | PCR programms based on [[Team:DTU-Denmark/Methods/PCR| standard PCR programm]] with annealing temperatures 52C for HAO and 56C for AMO and extension time 3:00 mins. | ||
| + | |||
| + | ===Colony PCR for Nir=== | ||
| + | In order to extract Nir from ''Pseudomonas'' we used primers which do not contain uracil and new PCR programm (touch-down PCR). Phusion polymerase was used. | ||
| + | |||
| + | Names of samples are as follows: | ||
| + | * 1,2 - primers 41a, 41b - Nir, part 1 | ||
| + | * 3,4 - primers 42a, 42b - Nir, part 2 | ||
| + | * 5,6 - primers 11a, 11b - all fragment | ||
| + | |||
| + | PCR programm (write it later when programm will be done) | ||
==Results== | ==Results== | ||
<hr/> | <hr/> | ||
| + | |||
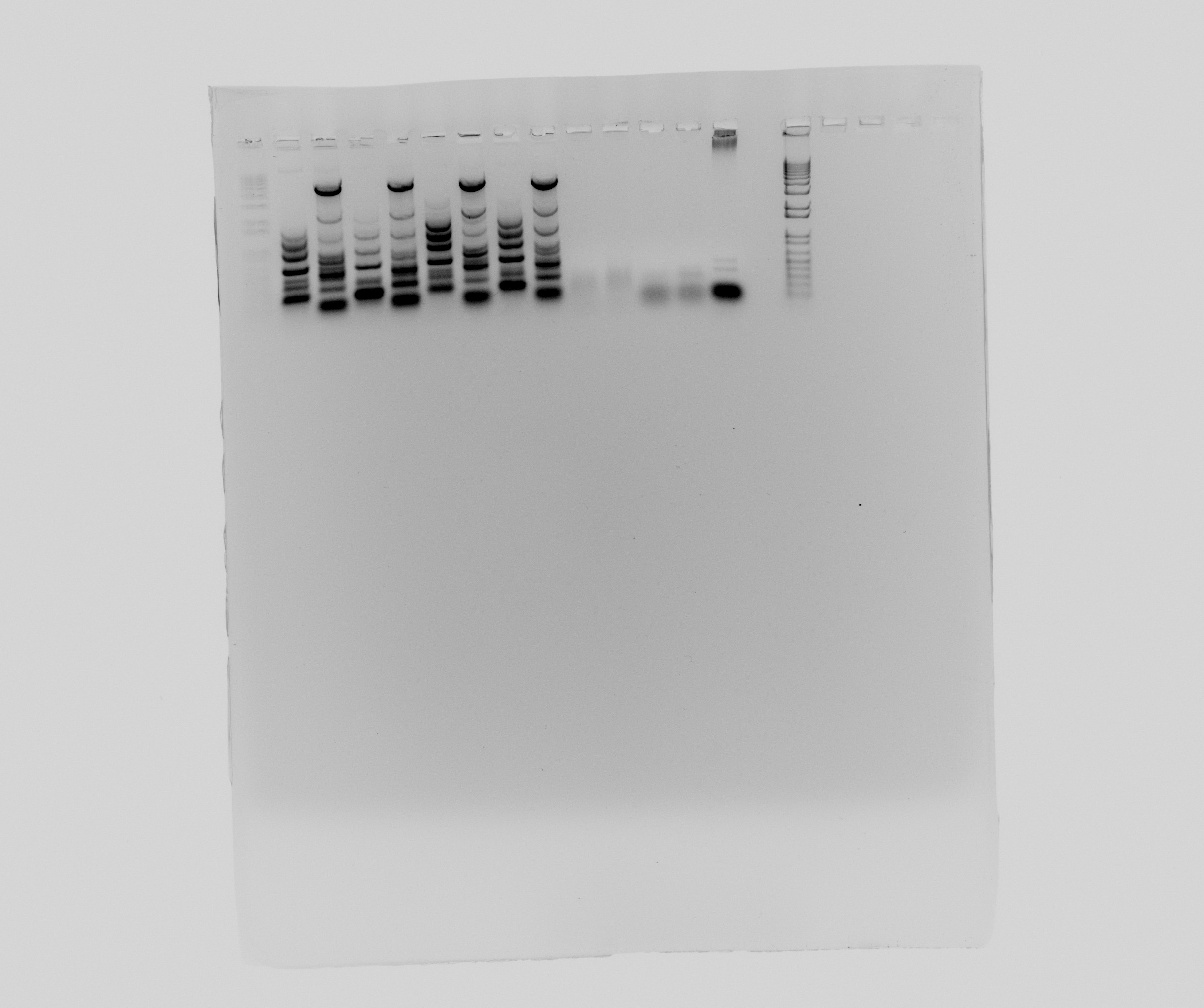
| + | ===Gel of PCR products from [[Team:DTU-Denmark/Notebook/28_July_2013| yesterday]]=== | ||
| + | |||
| + | * 1 - 1kb ladder | ||
| + | * 2 - Nir, part 1 | ||
| + | * 3 - Nir, part 2 | ||
| + | * 4 - Nir, part 1 | ||
| + | * 5 - Nir, part 2 | ||
| + | * 6 - Nir, part 1 | ||
| + | * 7 - Nir, part 2 | ||
| + | * 8 - Nir, part 1 | ||
| + | * 9 - Nir, part 2 | ||
| + | * 10 - pZE21 without promotor | ||
| + | * 11 - pZE21 without promotor | ||
| + | * 12 - HAO | ||
| + | * 13 - HAO | ||
| + | * 14 - negative | ||
| + | * 15 - 1 kb ladder | ||
| + | |||
| + | [[File:2013-07-29 nir pze hao.jpg|600px]] | ||
| + | |||
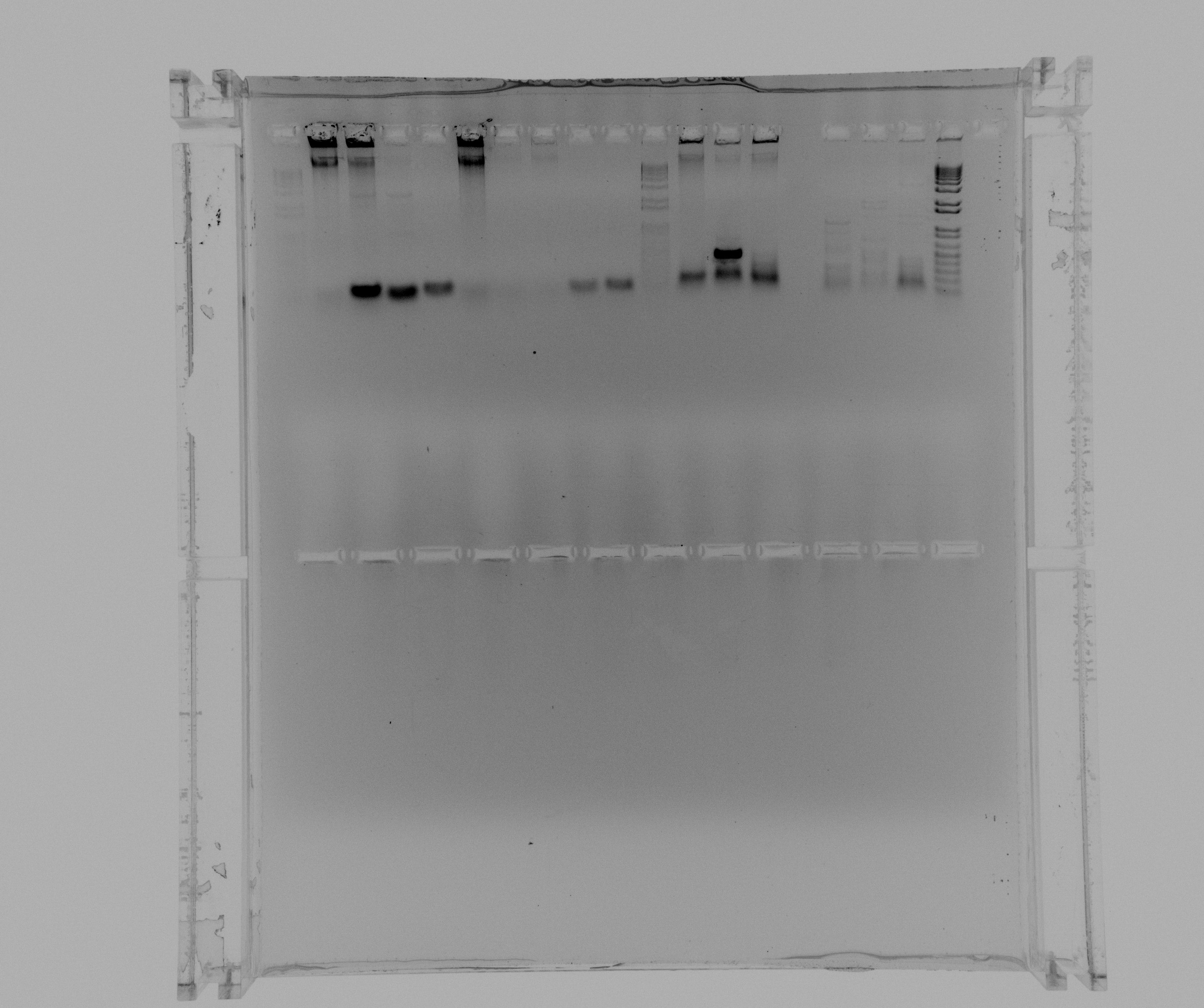
| + | ===Gel of PCR to extract Nir from ''Pseudomonas''=== | ||
| + | |||
| + | products of todays touchdown PCR | ||
| + | |||
| + | * 1 - 1 kb ladder | ||
| + | * 2 - Nir part 1 | ||
| + | * 3 - Nir part 1 | ||
| + | * 4 - Nir part 2 | ||
| + | * 5 - Nir part 2 | ||
| + | * 6 - broad band ladder | ||
| + | |||
| + | break | ||
| + | |||
| + | * 7 - 1 kb ladder | ||
| + | * 8 - Nir whole fragment | ||
| + | * 9 - Nir whole fragment | ||
| + | * 10 - broad band ladder | ||
| + | |||
| + | [[File:2013-07-29 nir new old.jpg|600px]] | ||
| + | |||
| + | |||
| + | ===Gel of todays colony PCR=== | ||
| + | |||
| + | Colony PCR to check for inserts in tranformants from USER reaction on the 24.07. | ||
| + | |||
| + | * 1 - 1 kb ladder | ||
| + | * 2 - AMO 1 | ||
| + | * 3 - AMO 2 | ||
| + | * 4 - AMO 3 | ||
| + | * 5 - AMO 4 | ||
| + | * 6 - AMO 5 | ||
| + | * 7 - AMO 6 | ||
| + | * 8 - AMO 7 | ||
| + | * 9 - AMO 8 | ||
| + | * 10 - AMO 9 | ||
| + | * 11 - 1 kb ladder | ||
| + | * 12 - HAO 1 | ||
| + | * 13 - HAO 2 | ||
| + | * 14 - HAO 3 | ||
| + | |||
| + | broken well | ||
| + | |||
| + | * 16 - HAO 4 | ||
| + | * 17 - HAO 5 | ||
| + | * 18 - HAO 6 | ||
| + | * 19 - 1 kb ladder | ||
| + | |||
| + | [[File:2013-07-29 colony amo hao.jpg|600px]] | ||
| + | |||
| + | faint bands of ~ 3000 kb can be seen in AMO colonies 2 and 3 | ||
| + | |||
| + | ===Genomic DNA isolation=== | ||
| + | Followed the protocol from the Qiagen: DNeasy Blood & Tissue Kit to isolate genomic DNA from the first PAO1 culture we got. Followed up by splitting the sample of purified DNA and treating one of them with HindIII which have no restriction sites within the Nir genes we are interested in. Then made PCR with primer set 11 on 65C and 4:30 extension. The reactions had either pure genomic DNA or HindIII treated and had a gradient of concentrations: 1, 3, 7, 10, 12, 15uL of DNA in place of MQ. | ||
==Conclusion== | ==Conclusion== | ||
<hr/> | <hr/> | ||
| + | |||
| + | PCR of Nir fragment gave a lot of unspecific products. New programm and new components for PCR are used to amplify it. | ||
| + | |||
| + | Innoculated ON cultures of AMO colony 2 and colony 3 for restriction analysis tomorrow. | ||
| + | |||
Navigate to the [[Team:DTU-Denmark/Notebook/28_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/30_July_2013|Next]] Entry | Navigate to the [[Team:DTU-Denmark/Notebook/28_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/30_July_2013|Next]] Entry | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Latest revision as of 20:44, 16 September 2013
29 July 2013
Contents |
Lab 208
Main purpose
- Colony PCR from Pseudomonas aeruginosa (PAO1) to isolate Nir.
- Colony PCR to check the presence of AMO and HAO in transformants E.coli.
Gel on yesterdays PCR Isolation of genomic DNA from PAO1
Who was in the lab
Kristian, Gosia, Henrike, Julia
Procedure
Colony PCR
Colony PCR was performed according to standard method in order to check if transformed cells which grew on medium contain desired insert in pZA21 plasmid. The colonies were chosen from agar plate to be checked. Names and numbers of colonies as well as plates names and primers are:
- HAO 1-6 in pZA21 USER, primers 28a, 28b
- AMO 1-9 in pZA21 USER, primers 29a, 29b
PCR programms based on standard PCR programm with annealing temperatures 52C for HAO and 56C for AMO and extension time 3:00 mins.
Colony PCR for Nir
In order to extract Nir from Pseudomonas we used primers which do not contain uracil and new PCR programm (touch-down PCR). Phusion polymerase was used.
Names of samples are as follows:
- 1,2 - primers 41a, 41b - Nir, part 1
- 3,4 - primers 42a, 42b - Nir, part 2
- 5,6 - primers 11a, 11b - all fragment
PCR programm (write it later when programm will be done)
Results
Gel of PCR products from yesterday
- 1 - 1kb ladder
- 2 - Nir, part 1
- 3 - Nir, part 2
- 4 - Nir, part 1
- 5 - Nir, part 2
- 6 - Nir, part 1
- 7 - Nir, part 2
- 8 - Nir, part 1
- 9 - Nir, part 2
- 10 - pZE21 without promotor
- 11 - pZE21 without promotor
- 12 - HAO
- 13 - HAO
- 14 - negative
- 15 - 1 kb ladder
Gel of PCR to extract Nir from Pseudomonas
products of todays touchdown PCR
- 1 - 1 kb ladder
- 2 - Nir part 1
- 3 - Nir part 1
- 4 - Nir part 2
- 5 - Nir part 2
- 6 - broad band ladder
break
- 7 - 1 kb ladder
- 8 - Nir whole fragment
- 9 - Nir whole fragment
- 10 - broad band ladder
Gel of todays colony PCR
Colony PCR to check for inserts in tranformants from USER reaction on the 24.07.
- 1 - 1 kb ladder
- 2 - AMO 1
- 3 - AMO 2
- 4 - AMO 3
- 5 - AMO 4
- 6 - AMO 5
- 7 - AMO 6
- 8 - AMO 7
- 9 - AMO 8
- 10 - AMO 9
- 11 - 1 kb ladder
- 12 - HAO 1
- 13 - HAO 2
- 14 - HAO 3
broken well
- 16 - HAO 4
- 17 - HAO 5
- 18 - HAO 6
- 19 - 1 kb ladder
faint bands of ~ 3000 kb can be seen in AMO colonies 2 and 3
Genomic DNA isolation
Followed the protocol from the Qiagen: DNeasy Blood & Tissue Kit to isolate genomic DNA from the first PAO1 culture we got. Followed up by splitting the sample of purified DNA and treating one of them with HindIII which have no restriction sites within the Nir genes we are interested in. Then made PCR with primer set 11 on 65C and 4:30 extension. The reactions had either pure genomic DNA or HindIII treated and had a gradient of concentrations: 1, 3, 7, 10, 12, 15uL of DNA in place of MQ.
Conclusion
PCR of Nir fragment gave a lot of unspecific products. New programm and new components for PCR are used to amplify it.
Innoculated ON cultures of AMO colony 2 and colony 3 for restriction analysis tomorrow.
Navigate to the Previous or the Next Entry
 "
"