Team:DTU-Denmark/Notebook/2 August 2013
From 2013.igem.org
(→lab 208) |
|||
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:DTU-Denmark/Templates/StartPage|2 August 2013}} | {{:Team:DTU-Denmark/Templates/StartPage|2 August 2013}} | ||
| - | + | Navigate to the [[Team:DTU-Denmark/Notebook/1_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/3_August_2013|Next]] Entry | |
| - | = | + | =Lab 208= |
<hr/> | <hr/> | ||
==Main purpose== | ==Main purpose== | ||
| Line 47: | Line 47: | ||
===PCR on Nir fragment 2 with GC-buffer=== | ===PCR on Nir fragment 2 with GC-buffer=== | ||
| - | Same procedure as | + | Same procedure as [[Team:DTU-Denmark/Notebook/1_August_2013#PCR_on_Nir_and_single_fragment_Nir|yesterday's PCR]] just with GC-buffer instead of HF buffer. |
==Results== | ==Results== | ||
| Line 64: | Line 64: | ||
| - | Gel with Nir as single gene fragments and PAO1 from environmental lab as template: | + | Gel with Nir as single gene fragments and PAO1 from environmental lab as template (only Nir E samples): |
*1kb ladder | *1kb ladder | ||
*Nir fragment 2, expected length = 5000 | *Nir fragment 2, expected length = 5000 | ||
| Line 101: | Line 101: | ||
==Conclusion== | ==Conclusion== | ||
<hr/> | <hr/> | ||
| + | We have something that looks like Nir fragment 1 and further experiments will verify that. We speculate that the high GC-content can be the reason why we have had so many PCR problems with Nir and therefore we are doing the next PCR on Nir 2 with GC-buffer. | ||
| + | |||
| + | |||
Navigate to the [[Team:DTU-Denmark/Notebook/1_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/3_August_2013|Next]] Entry | Navigate to the [[Team:DTU-Denmark/Notebook/1_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/3_August_2013|Next]] Entry | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Latest revision as of 20:45, 16 September 2013
2 August 2013
Contents |
Lab 208
Main purpose
- Gel purification of Cyc and Nir
- USER reaction for making His-tag on cycAX construct
- PCR of Nir, His-Tag Sec, His-Tag TAT
- Miniprep re-purification with columns
- Inoculation of samples for midi prep
Who was in the lab
Gosia, Kristian, Natalia, Julia
Procedure
Gel purification
- Gel purification was performed according to protocol included in QIAEX, Gel Extraction Kit.
USER reaction
- USER reaction was performed according to standard protocol SOP1. We use DNA linear fragment containing pZA21 bacbone,cycAX with His-Tag sequence.
PCR
- PCR according to standard PCR protocol.
Samples names:
- 1,2 - Nir 48 (primers 48a, 48b) on template of Nir (too long fragment amplified at the beginning of work with Nir PCR)
- 3,4 - Sec His-Tag, primers 19a, 19b; template: Sec 2
- 5,6 - Tat His-Tag, primers 20a, 20b; template: TAT 2 1
- 7,8 - Tat His-Tag, primers 20a, 20b; template: TAT 3 2
- 9, 10 - Tat His-Tag, primers 20a, 20b; template: TAT 3 1a
- 11, 12, 13 - negative controls as follows for: Tat His-Tag, Nir, Sec His-Tag
Program for samples:
- 1,2,12 - Program for Nir on Eppendorf machine
- 3,4,13 - 10 cycles 58.5 C annealing, 1 min elongation and 25 cycles on 67 C annealing
- 5-11 - 10 cycles on 54.5 C annealing and 25 cycles on 64 C annealing
Purification of plasmids after ethanol purification
According to protocol attached to the DNA purification kit, QIAGEN
Inoculation
For tomorrows mini prep 5 samples were prepared (all in shaker at 37 C, last line). To 10 ml of LB with kanamycin 100 uL of inoculum was added (from previous liquid culture)
PCR on Nir fragment 2 with GC-buffer
Same procedure as yesterday's PCR just with GC-buffer instead of HF buffer.
Results
Purification gel for yesterdays PCR on on cycAX with His-tag and AMO with USER primers:
- 1kb ladder
- AMO
- AMO
- AMO
- cycAX his-tag
- cycAX his-tag
- cycAX his-tag
- 1kb ladder
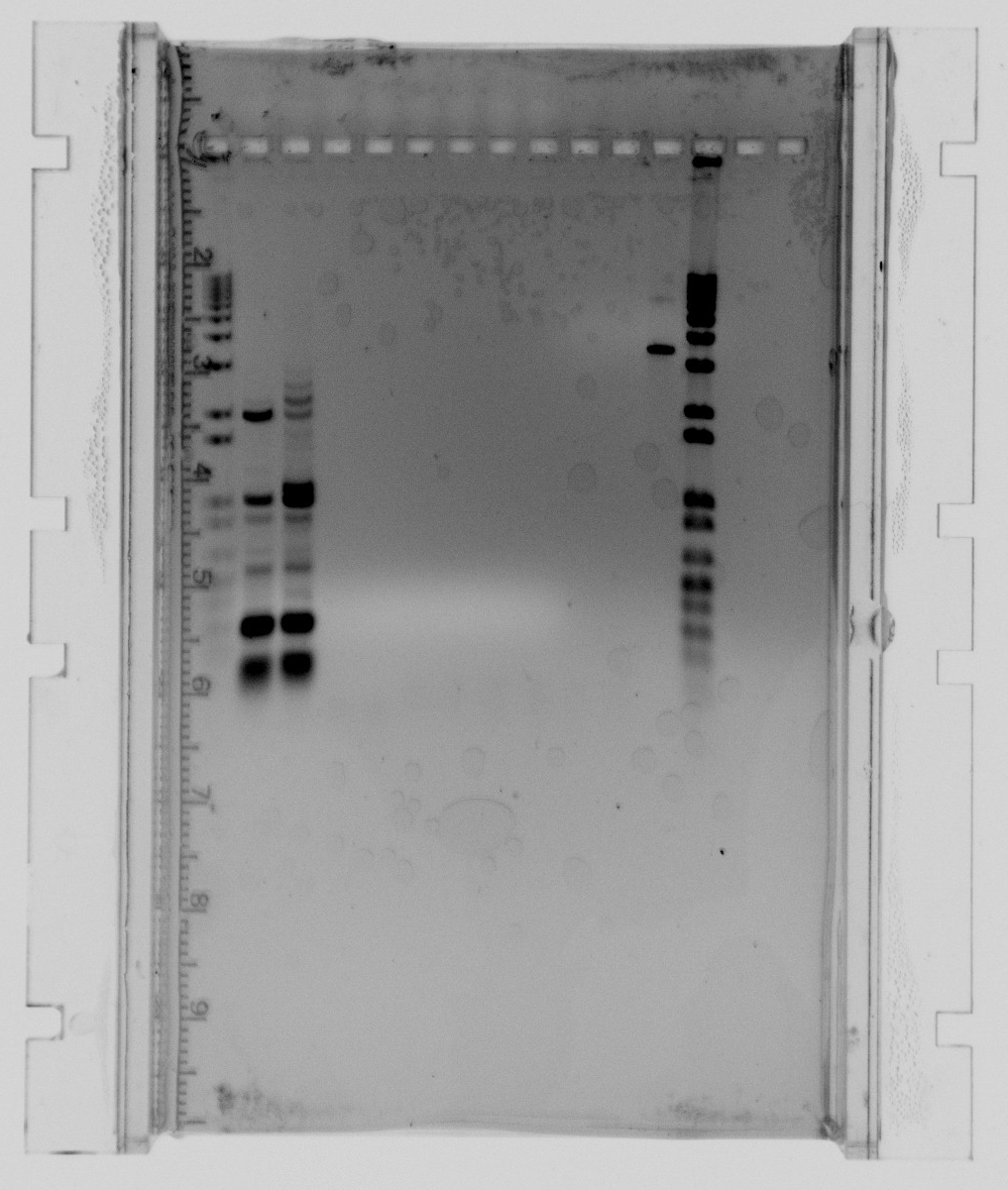
Gel with Nir as single gene fragments and PAO1 from environmental lab as template (only Nir E samples):
- 1kb ladder
- Nir fragment 2, expected length = 5000
- Nir fragment 2, expected length = 5000
- Nir fragment 1, expected length = 4100
- Nir fragment 1, expected length = 4100
- Nir whole fragment with KD primers, expected length = 9100
- Nir whole fragment with KD primers, expected length = 9100
- NirN, expected length = 1700
- NirN, expected length = 1700
- NirM+S, expected length = 2300
- NirM+S, expected length = 2300
- NirG, expected length = 650
- NirG, expected length = 650
- Neg. control
- 1kb ladder
Gel on todays PCR with his-tag and verification of Nir:
- 1kb ladder
- Nir previous purified tested with primers for NirG expected length = 650
- Nir previous purified tested with primers for NirG expected length = 650
- Sec His-Tag
- Sec His-Tag
- TAT His-Tag
- TAT His-Tag
- TAT His-Tag
- TAT His-Tag
- TAT His-Tag
- TAT His-Tag
- 1kb ladder
Conclusion
We have something that looks like Nir fragment 1 and further experiments will verify that. We speculate that the high GC-content can be the reason why we have had so many PCR problems with Nir and therefore we are doing the next PCR on Nir 2 with GC-buffer.
Navigate to the Previous or the Next Entry
 "
"