Team:DTU-Denmark/Codon Optimization
From 2013.igem.org
| (8 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:DTU-Denmark/Templates/StartPage|}} | {{:Team:DTU-Denmark/Templates/StartPage|}} | ||
| + | __TOC__ | ||
| + | == Abstract and Methods == | ||
| - | + | The purpose of this work is to determine whether we should codon optimize the genes from ''Nitrosomonas europaea'' and ''Pseudomonas aeruginosa'' that we are expressing in ''E. coli''. We find that the heterologous expression of these genes is such that we do not need to codon optimize. | |
| - | + | ''E. coli'' has a different tRNA pool than both ''N. europaea'' and ''P. aeruginosa'' do. When genes from these organisms are expressed in ''E. coli'', they may have a very inefficient translation efficiency if the available tRNA pools are very different. To determine the impact of this difference, we calculated the tRNA adaptation index (tAI). The tAI for a gene is a measure of the availability of the tRNAs that serve each codon in the gene. A high tAI for a gene means the gene incorporates more rare tRNAs (tRNAs that do not occur frequently in the genome sequence) than a gene with lower tAI would. A high tAI also indicates that the gene has been optimized for high expression levels. | |
| - | + | Using tRNAscan-SE, we determined the tRNA codon copy numbers for ''E. coli'', ''N. europaea'' and ''P. aeruginosa''. Then, we calculated the tAI for each organism using a script provided by Chris Workman (unpublished). Finally, we calculated the tAI of heterologous expression of our genes of interest from ''N. europaea'' and ''P. aeruginosa'' in ''E. coli'' using the same script, but with the ''E. coli'' codon table in place of the native codon table. The tAI graphs were plotted using the R package ggplot2. | |
| - | + | ||
| - | Using tRNAscan-SE | + | |
== Results == | == Results == | ||
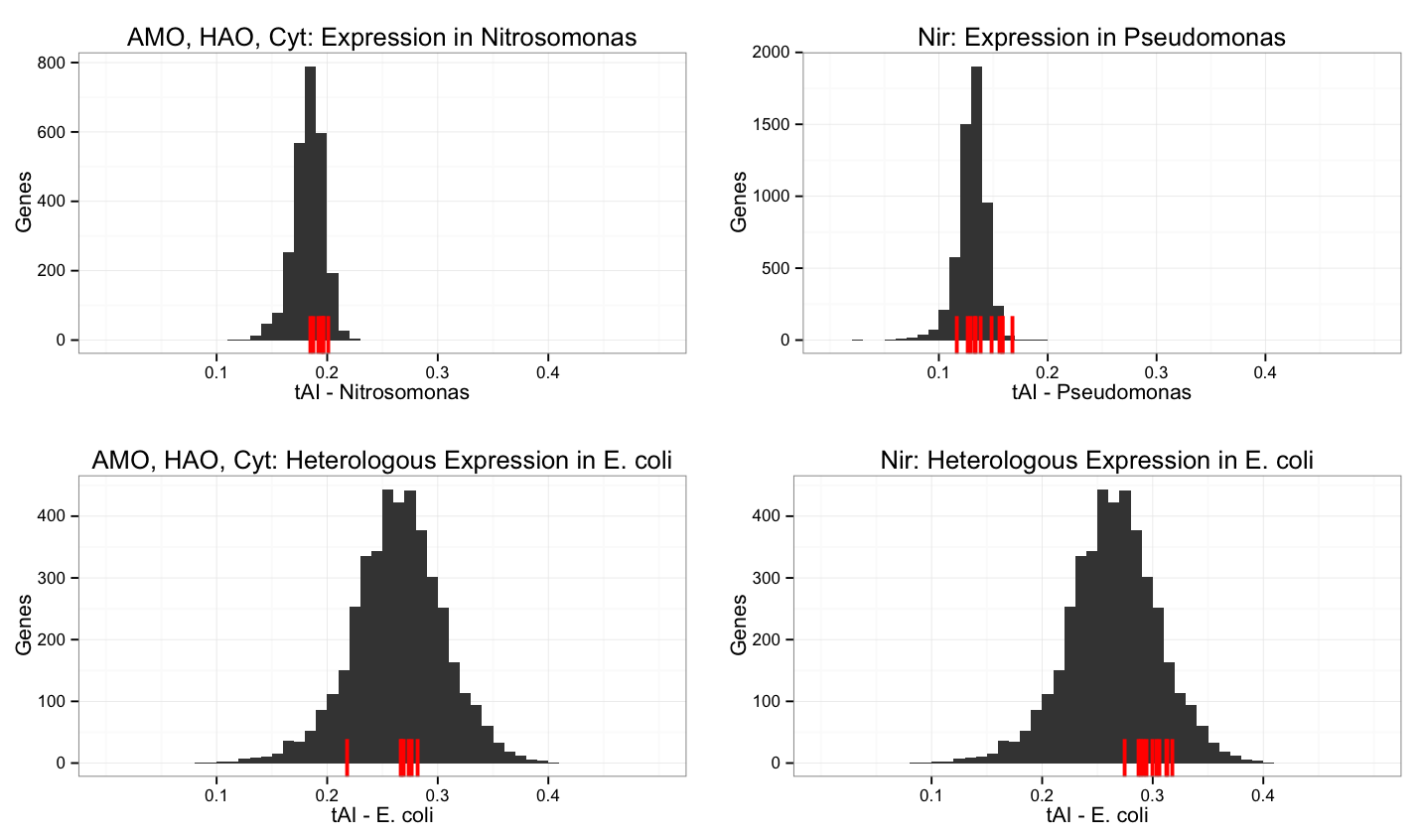
| - | + | Graphs are shown for genes from ''Nitrosomonas europaea'' (left) and ''Pseudomonas aeruginosa'' (right). The top two graphs shows tAI levels over these organisms, with our genes of interest marked in red (AMO, HAO and cytochromes for ''N. europaea'' and Nir for ''P. aeruginosa''), and the bottom two graphs show heterologous expression of the same genes of interest in ''E. coli''. The grey area shows the distribution of tAI values over all genes in the organism specified. Click for a larger image. | |
| - | + | ||
| - | + | [[File:Dtu codon opt.png|600px]] | |
| - | [[File:Dtu codon opt | + | |
| - | == Conclusions == | + | == Conclusions and Discussion == |
Without codon optimization, the genes that we are interested in have tAI values close to the mean for ''E. coli''. This is sufficient for our purposes, and codon optimization is not necessary. If we wanted to express these genes at a higher rate, there is room to codon optimize, however, since many of these genes code for membrane proteins, we are concerned about expression at too high a level that will lead to membrane stress in ''E. coli''. | Without codon optimization, the genes that we are interested in have tAI values close to the mean for ''E. coli''. This is sufficient for our purposes, and codon optimization is not necessary. If we wanted to express these genes at a higher rate, there is room to codon optimize, however, since many of these genes code for membrane proteins, we are concerned about expression at too high a level that will lead to membrane stress in ''E. coli''. | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Latest revision as of 18:03, 4 October 2013
Contents |
Abstract and Methods
The purpose of this work is to determine whether we should codon optimize the genes from Nitrosomonas europaea and Pseudomonas aeruginosa that we are expressing in E. coli. We find that the heterologous expression of these genes is such that we do not need to codon optimize.
E. coli has a different tRNA pool than both N. europaea and P. aeruginosa do. When genes from these organisms are expressed in E. coli, they may have a very inefficient translation efficiency if the available tRNA pools are very different. To determine the impact of this difference, we calculated the tRNA adaptation index (tAI). The tAI for a gene is a measure of the availability of the tRNAs that serve each codon in the gene. A high tAI for a gene means the gene incorporates more rare tRNAs (tRNAs that do not occur frequently in the genome sequence) than a gene with lower tAI would. A high tAI also indicates that the gene has been optimized for high expression levels.
Using tRNAscan-SE, we determined the tRNA codon copy numbers for E. coli, N. europaea and P. aeruginosa. Then, we calculated the tAI for each organism using a script provided by Chris Workman (unpublished). Finally, we calculated the tAI of heterologous expression of our genes of interest from N. europaea and P. aeruginosa in E. coli using the same script, but with the E. coli codon table in place of the native codon table. The tAI graphs were plotted using the R package ggplot2.
Results
Graphs are shown for genes from Nitrosomonas europaea (left) and Pseudomonas aeruginosa (right). The top two graphs shows tAI levels over these organisms, with our genes of interest marked in red (AMO, HAO and cytochromes for N. europaea and Nir for P. aeruginosa), and the bottom two graphs show heterologous expression of the same genes of interest in E. coli. The grey area shows the distribution of tAI values over all genes in the organism specified. Click for a larger image.
Conclusions and Discussion
Without codon optimization, the genes that we are interested in have tAI values close to the mean for E. coli. This is sufficient for our purposes, and codon optimization is not necessary. If we wanted to express these genes at a higher rate, there is room to codon optimize, however, since many of these genes code for membrane proteins, we are concerned about expression at too high a level that will lead to membrane stress in E. coli.
 "
"