Jsher
From 2013.igem.org
(Difference between revisions)
(→August 29, 2013) |
|||
| (7 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<div id="header">{{Template:Team:Yale2013/Templates/Header}}</div> | <div id="header">{{Template:Team:Yale2013/Templates/Header}}</div> | ||
| + | <br> | ||
| + | '''[https://2013.igem.org/Team:Yale/Notebook Return to Lab Notebooks]'''<br> | ||
| + | |||
| + | ==September 27, 2013== | ||
| + | *Tested 36, 64, 65, 66 from frozen stocks grown overnight in LB, NR, ATC, Arabinose, Kan | ||
| + | **Spun down 1ml of the cells, washed in PBS once and put 150ul of cells + PBS into 750ul of PBS | ||
| + | **FACS sorted again using PE-Texas red | ||
| + | **Sorted all three tubes according to a gate that had 15 of the baseline (which was 38 or our cells with our plasmid) | ||
| + | **KO had 26, RBS had 65, while All had 98 | ||
| + | ***We also set a higher gate and sorted the all sample again this time only getting the top 0.1% which was 7/10,000 | ||
| + | *I started cultures of all of these and will plate read this weekend | ||
| + | |||
| + | ==September 20, 2013== | ||
| + | *Made new LB and LB agar--for Spec plates later | ||
| + | *Going to test the plLacO clones 68-71, and use 38 as a control, going to put in LB, NR, IPTG, Arabinose, Kan | ||
| + | **15ml LB, 7.5ul NR, 15ul IPTG, 75ul AR, 15ul Kan | ||
| + | |||
| + | *Start cultures of 64, 65, 66 in LB Kan (Kan is 1000x) for MAGE. These all have our plasmid already | ||
| + | 64-->KO oligos, 65---> RBS oligos, 66---all Oligos | ||
| + | |||
| + | *Start 2 cultures of #1 in LB. These don't have our plasmid and would be for later transformations with fixed promoter thing---One is for KO oligos, one for RBS and one for all | ||
| + | |||
| + | |||
| + | |||
| + | ==September 16, 2013== | ||
| + | *My redoing the biobrick did not work, so we just submitted 5.35ul of PCT1 which was at 45.5ng/ul in 10ul of nuc free H2O | ||
| + | **PCT1 is in the plasmid box and in the frozen stock and is a white colony | ||
| + | |||
| + | |||
| + | ==September 15, 2013== | ||
| + | *Running the PCRs of the PCT gene and backbone (2 versions) on a gel--working on making our biobrick | ||
| + | **Gel purified and nanodroped--the DNA tubes are is in the cold block in our -20 | ||
| + | **Set up and ran Gibson assemblies | ||
| + | **In PCR 3A on old PCR machines | ||
| + | |||
| + | *Looked at the tetR specR plates and there are 5 or so colonies on the negative control and many more on the positive | ||
| + | **Some sort of contamination present, so will put in the 4 degrees but probably don't want to use | ||
| + | |||
| + | *There is a gibson assembly of plLacO and our construct done---need to transform into EcNR2 and plate on Kan | ||
| + | |||
| + | |||
| + | ==September 11, 2013== | ||
| + | *Setup plate reader control to test all 4 of the positive plLacO constructs | ||
| + | **Growing frozen stock #1, #38, #68-71 in LB NR AR IPTG and as a control growing #38 and 68 in LB NR AR ATC, and 1 in LB NR | ||
| + | *NR is 2000X, ATC is 1000X, Arabinose is 100X, IPTG is 1000X | ||
| + | |||
| + | *Start big culture for PLA extraction | ||
| + | **Going to start a 5ml culture of frozen stock number 38 with 5ul of Kan and let grow overnight | ||
| + | **Also, making a 1L of LB in a 4000ml flask which will be autoclaved | ||
| + | **Then tomorrow need to add the 5ml of confluent cells to the 4L flask and let grow to mid log | ||
| + | ***Then add inducers (ATC and arabinose), then let grow overnight | ||
| + | |||
| + | |||
| + | *Redoing PCT amplifications to put on biobrick because we need to do this ASAP | ||
| + | **Going to amplify PCT off of our entire construct (164,165 and 168 and 169) | ||
| + | **Apparently Tim didn't amplify up the biobrick to give it homology to PCT, which we should have done so I just ordered 4 new primers which have the stop codon and amplify up the backbone so the assembly should work a lot better | ||
| + | **bio_PCT_R gggaacctttctcatctctagaagcggccgcgaa | ||
| + | **bio_PCT_F gaaatgaagtccTAAtactagtagcggccgtgcagtccgg | ||
| + | **PCT_Bio_F cggccgcttctagagatgagaaaggttcccattattaccgc | ||
| + | **PCT_Bio_R cggccgctactagtaTTAggacttcatttccttcagacccatt | ||
| + | |||
| + | ==September 9, 2013== | ||
| + | *Here is a graph of the data from switching the promoters | ||
| + | **Does not appear to have worked so probably some of the old plasmid make it through and thus they are really the same---need to DPN1 digest before we transform | ||
| + | [[File:EcNR2 switching promoters.jpg|500px]] | ||
| + | *This is the plates from the second attempt at the specR TetR transformation. | ||
| + | **Getting the feeling that these spec plates are bad so going to make new ones and try again | ||
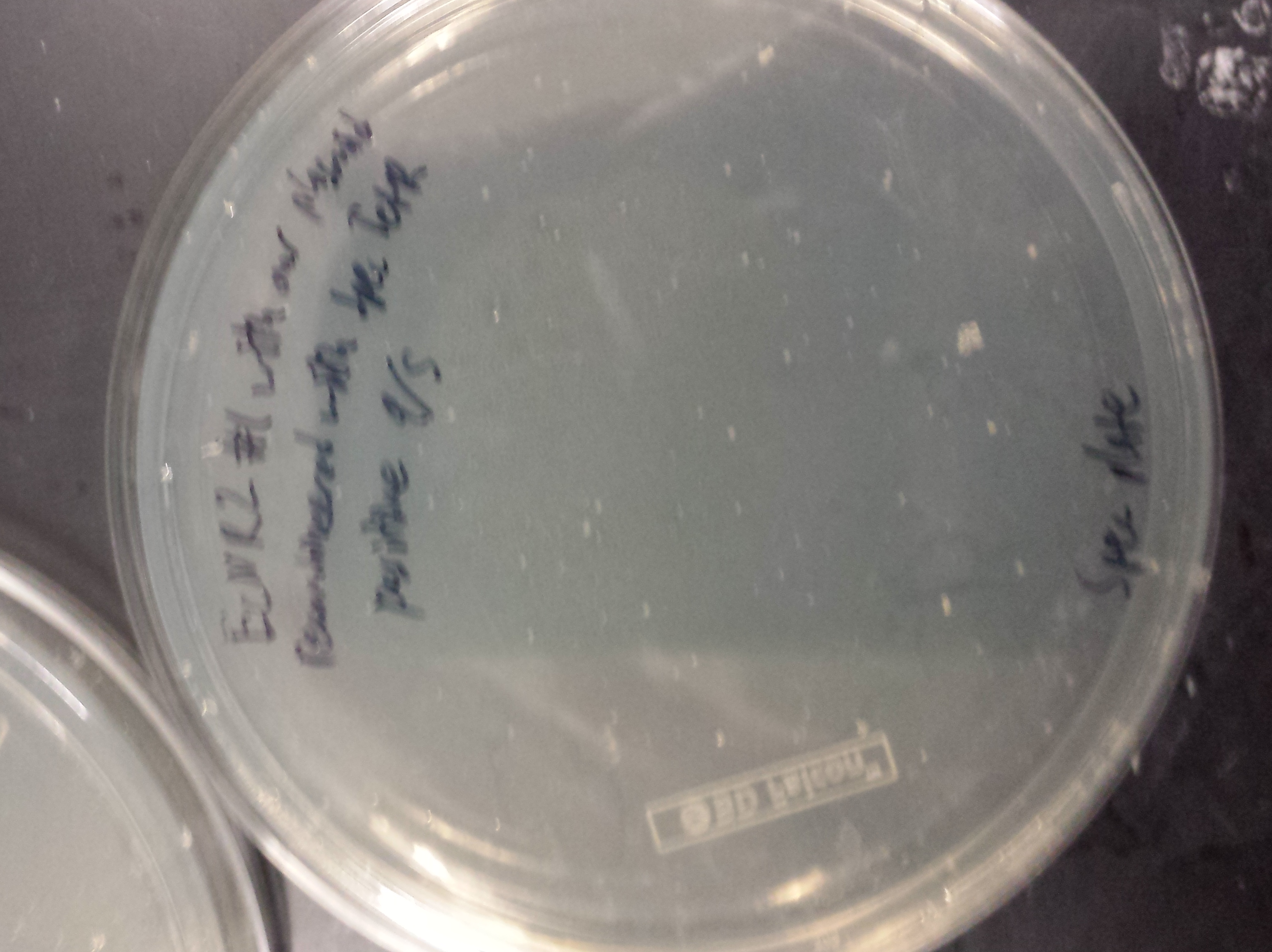
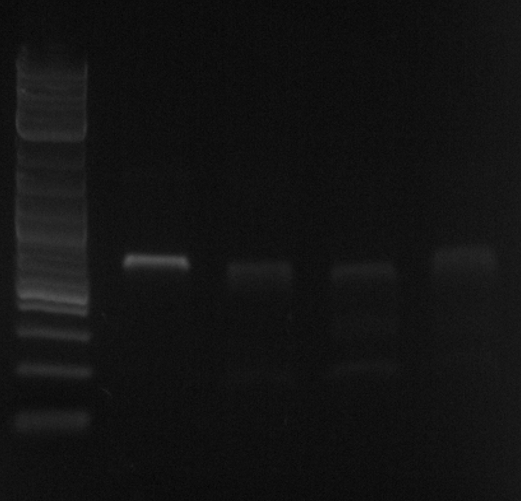
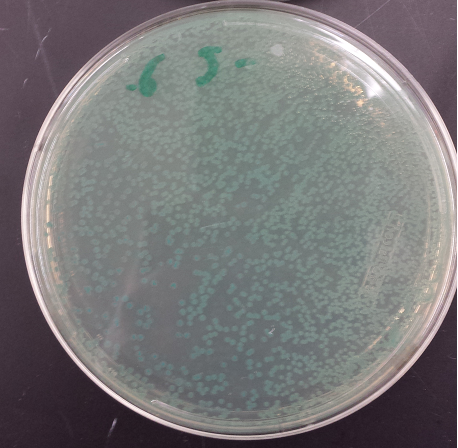
| + | [[File:Spectrial1.png|300px]] | ||
| + | |||
| + | ==September 7, 2013== | ||
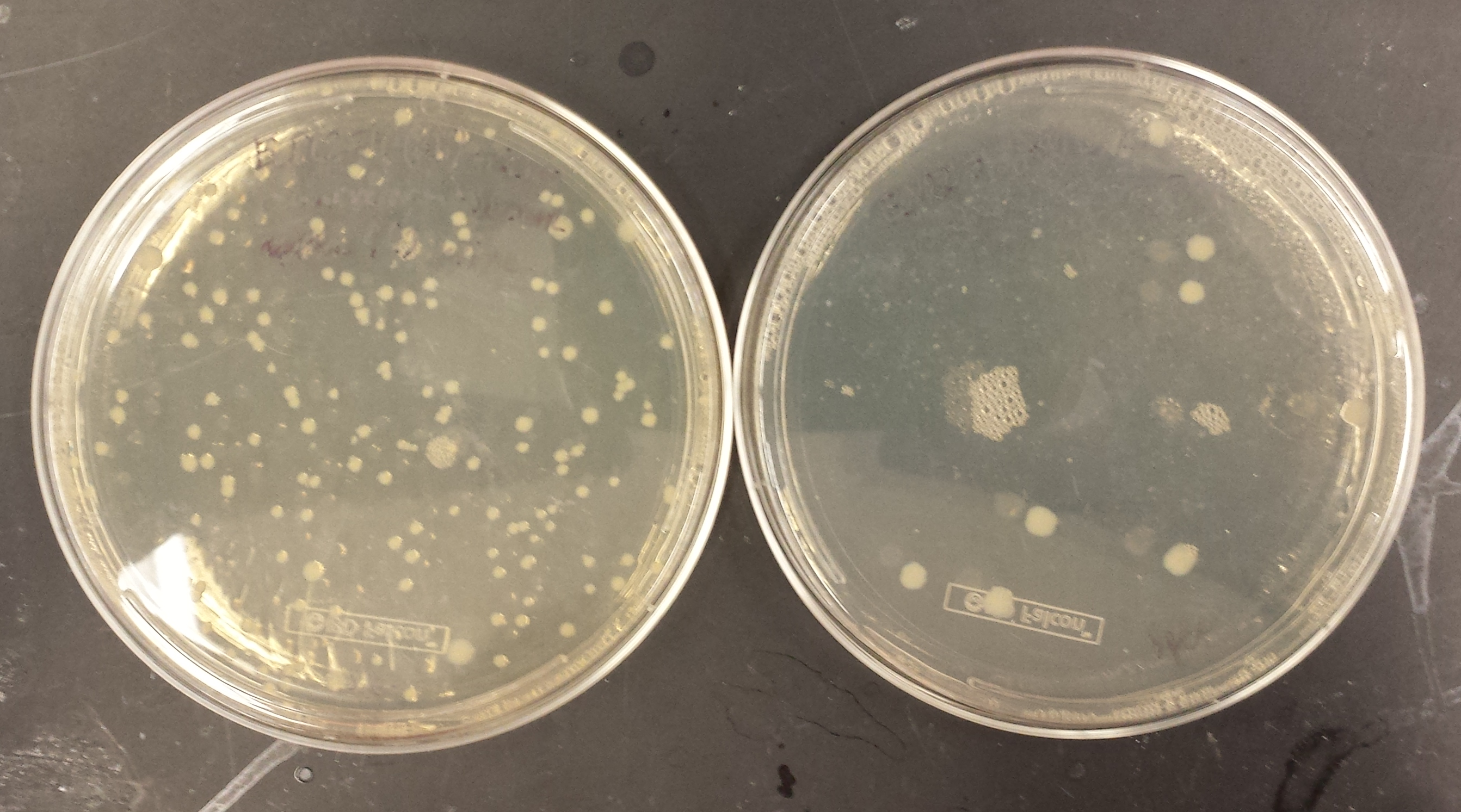
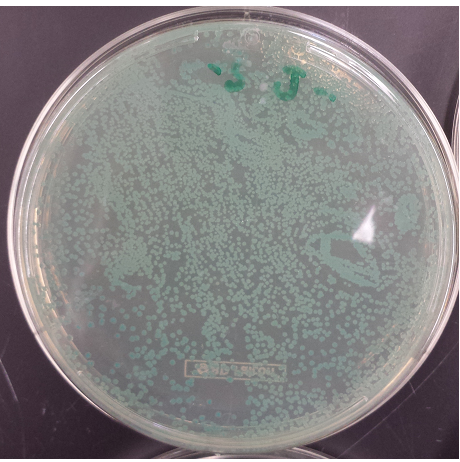
| + | [[File:Spectrial3.png|300px]] | ||
| + | [[File:Spectrial2.png|300px]] | ||
| + | *Picked 46 colonies into 96 well plate---#1 is negative control (EcNR2) and replacing #48 with the DsDNA from Jamin | ||
| + | ***primers 172, 173--30 second extension, 65 degrees | ||
| + | ***Realized I used the original specR which means homolgoy is only 58 degrees to that so the "positive" control might not work properly | ||
| + | **Run ladder, A1, B1.....ladder in row 1 | ||
| + | **Run ladder, C1, D1.....ladder in row 2 | ||
| + | [[File:Farren 2013-09-07 16hr 08min.jpg|400px]] | ||
| + | |||
| + | *So I grew the Biobricks in Spec by accident and none grew (good sign really since they actually have Cat resistance not spec) | ||
| + | **So regrew all 16 in LB, and will attempt to PCR tomorrow | ||
| + | |||
| + | *Andrew is drop dialyzed and is transforming the gibson assembly from yesterday into EcNR2 #1, and will plate on Kan plates in 1 hour. | ||
| + | |||
| + | |||
| + | ==September 6, 2013== | ||
| + | *Checked spec plates, and there appears to be some colonies, but will let grow overnight to get a clearer picture | ||
| + | |||
| + | *Going to attempt to gibson assemble plLacO promoter and our plasmid minus the plTetO promoter | ||
| + | *Using 3.62ul of the 39.6ng/ul Backbone, with either 1ul or 3ul of the 27.9ng/ul Promoter | ||
| + | **[https://docs.google.com/a/yale.edu/spreadsheet/ccc?key=0AmKfFDrNoTUadFp1d2dOMEFiUmpKOGFnc0l0RjNXLXc#gid=0 Here are amounts ] | ||
| + | **Going to transform into EcNR2 #1 | ||
| + | |||
| + | *Spun down 50ml of the overnight grown #38 with ATC and arabinose cultures | ||
| + | **Andrew grew in incubator (non-shaking) in a 100ml bottle | ||
| + | *Ben: Bacterial pellets were washed with acetone for 20 minutes | ||
| + | |||
| + | |||
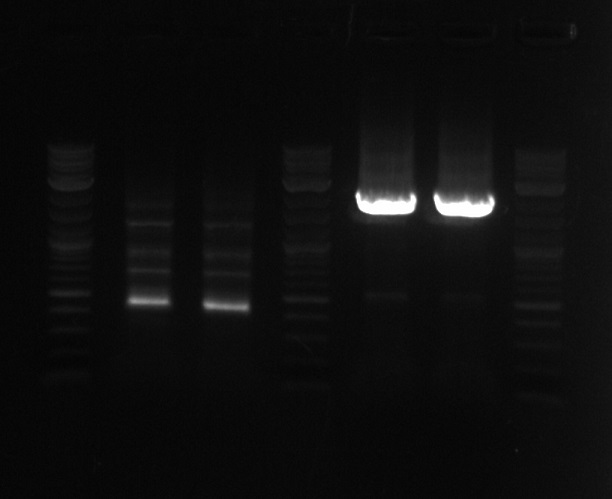
| + | *Growing up all 16 possible cultures which could have PCT on a biobrick---growing them in 1ml LB with 1ul spec | ||
| + | **Will need to PCR test all of them tomorrow--same primers and such Andrew did yesterday | ||
| + | **Also going to need a positive control with the DNA that was used the gibson assembly of the biobrick---try to amplify up the PCT gene with homology to the Biobrick | ||
| + | |||
| + | |||
| + | *Designed and ordered 3 primers to test for spec or TetR | ||
| + | **forward primer: GCATTTATCAGGGTTATGTCCACTGG | ||
| + | **Reverse for tetR: AAGGCGACGTGCGTCCT 1kb | ||
| + | **Reverse for Spec: GGCCAGCAACGTCGGTTC 0.5kb | ||
| + | |||
| + | *Order screening primers so this locus at the genome and look for either spec which is 1.2kb or tetR which is 600bp | ||
| + | **F:GCATTTATCAGGGTTATGTCCACTGG | ||
| + | **R:AGTTTAAATATGGCTGGTTCTCGCAG | ||
| + | |||
| + | ==September 3, 2013== | ||
| + | *PCR PCT out of Biobrick | ||
| + | **Going to try and PCR out our gene---using primers 27 and 30 to make about a 1kb fragment, 58 degrees, 30 second extension | ||
| + | **PCR 1:PCT1, 2:PCT4, 3:PCT5, 4:PCT8 | ||
| + | ***PCT1 and PCT 5 were white while PCT 4 and 8 were red | ||
| + | *Redo Spec PCR with Jaymen's plasmid | ||
| + | **PCR 5---primers 172, 173--45 second extension, 58 degress | ||
| + | *Ran a gel: ladder, PCR5, ladder, PCR 1-4 ladder | ||
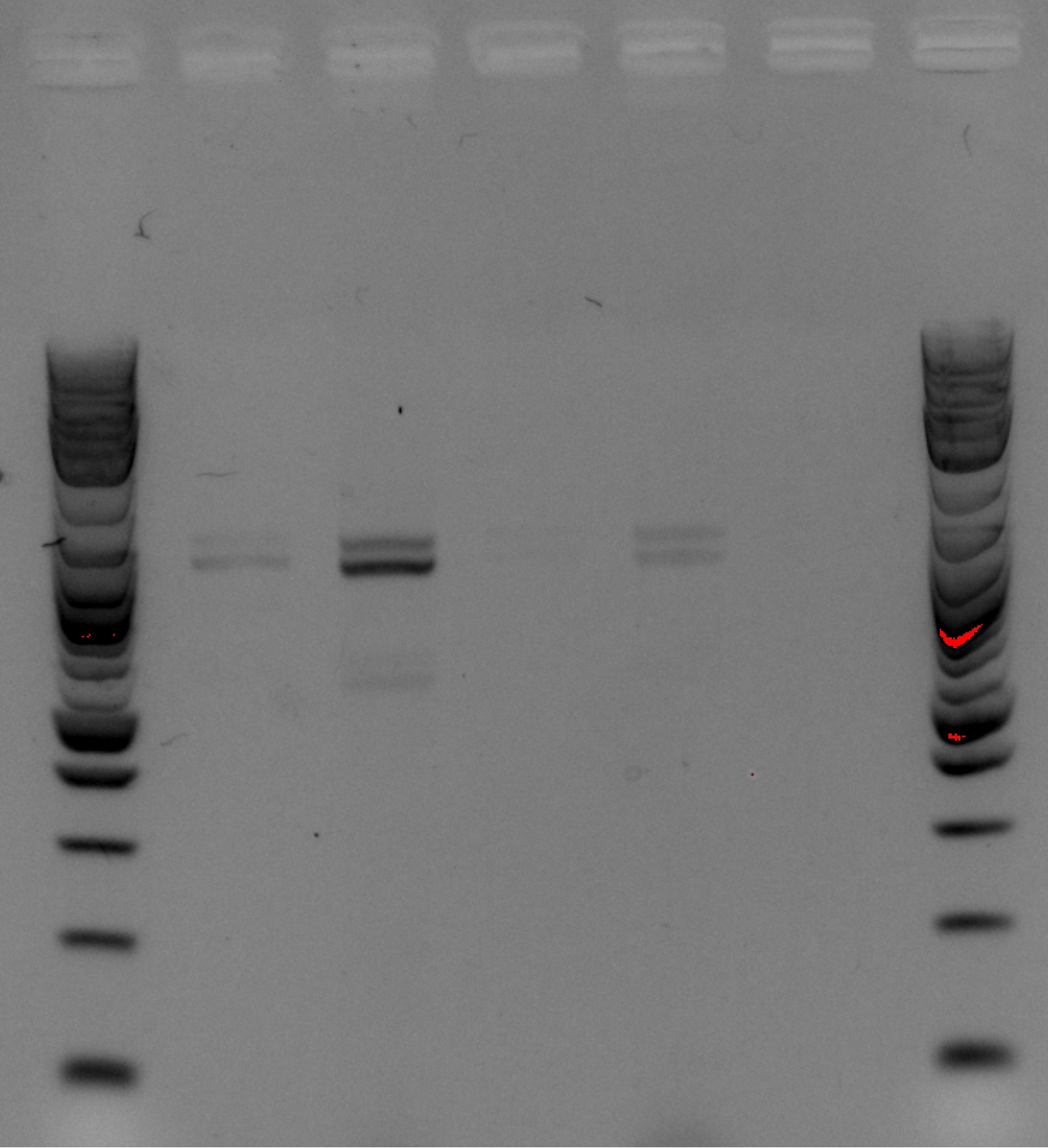
| + | [[File:Gel9-3.png|600px]] | ||
| + | *Gel purified and nanodroped the SpecR with homology to TetR (22.2ng/ul)---in DsDNA2 box | ||
| + | |||
| + | *Start working on replacing PlTetO with plLacO | ||
| + | *PCR PZE21 6185bp, primers 178,179 58 degrees for 1:45sec extension (6) | ||
| + | *PCR plLacO 136bp, primers 180 and 181, 58 degres for 15 second extension (7) | ||
| + | |||
| + | |||
| + | |||
| + | ==September 1, 2013== | ||
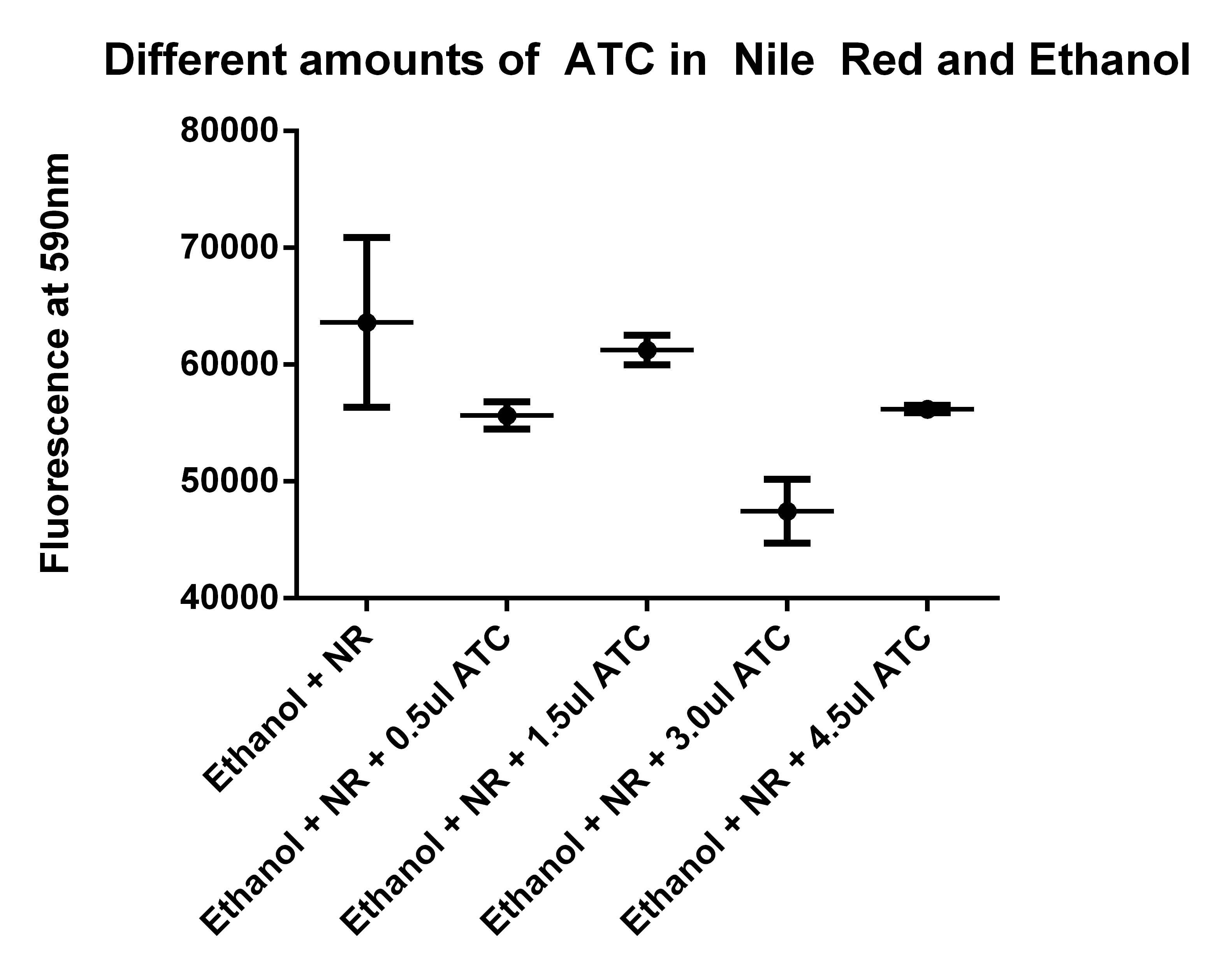
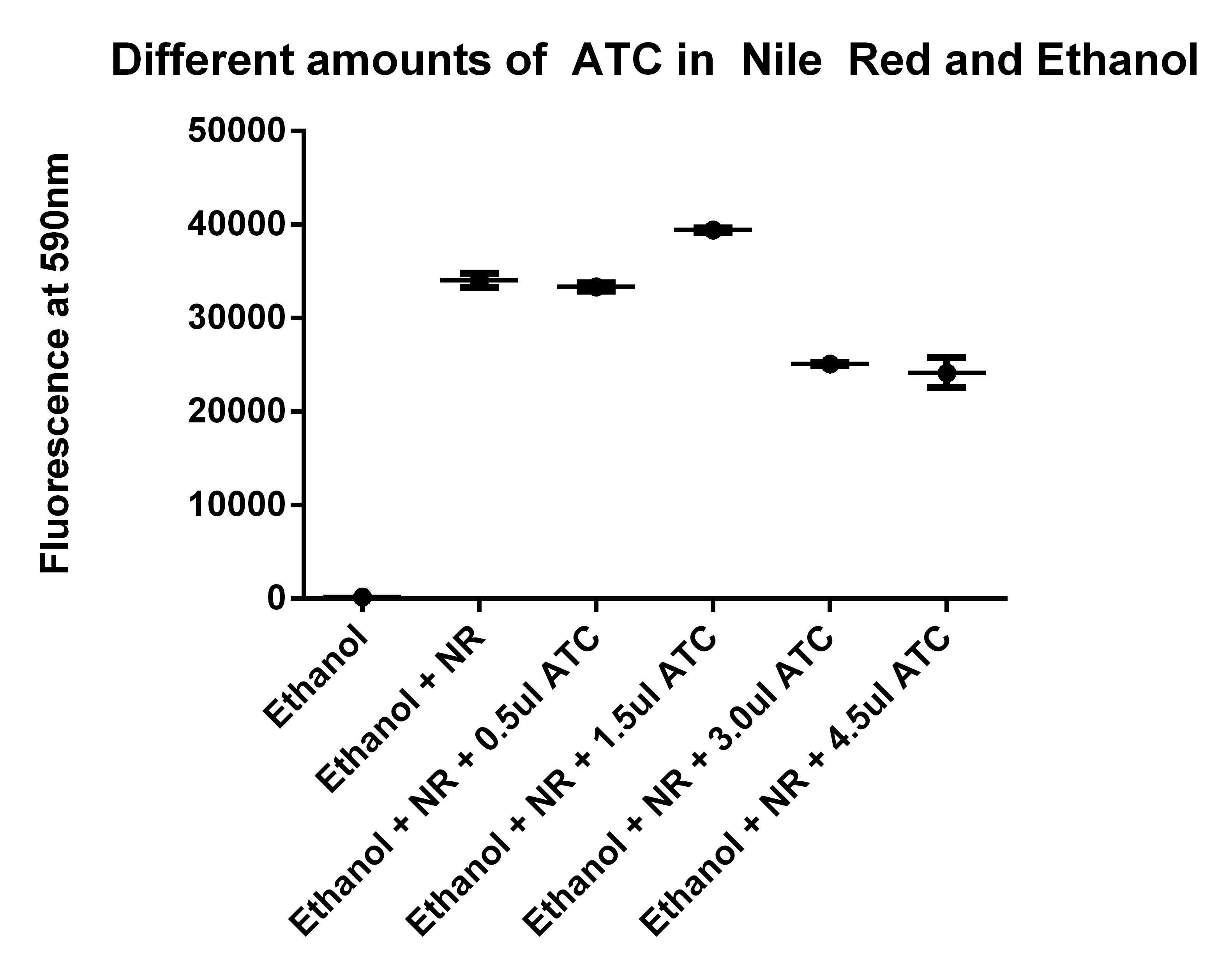
| + | *Going to test Ethanol + NR with different amounts of ATC | ||
| + | [[File:NR and ATC9-1.jpg|400px]] | ||
| + | |||
| + | *Spun and resuspended cells from yesterday | ||
| + | **Spun down 1ml of the confluent cells, resuspended in LB or LB + NR, and then spun down again and resuspended in LB or LB + NR | ||
| + | **Put back in the incubator for 1 hour (starting at 2pm), and then will test on plate reader | ||
| + | **Will test #38 and #1 as a control | ||
| + | #No NR, AR, or ATC | ||
| + | #AR and ATC, will spin down tomorrow and resuspended in LB | ||
| + | #AR and ATC, will spin down tomorrow and resuspend in Nile Red LB | ||
| + | #NR, AR, and ATC will spin down tomorrow and resuspended in Nile Red LB | ||
| + | #NR, AR, and ATC | ||
| + | *Results | ||
| + | [[File:Nile red Strain.jpg|400px]] | ||
| + | |||
| + | |||
| + | *It appears that the Nile red might have gone bad, so I made new Nile red and used it in these new samples | ||
| + | *Going to test letting the cells grow overnight in ATC and Arabinose and staining the next day wth Nile red | ||
| + | **Will test #38 and #1 as a control | ||
| + | #No NR, AR, or ATC | ||
| + | #AR and ATC, will spin down tomorrow and resuspended in LB | ||
| + | #AR and ATC, will spin down tomorrow and resuspend in Nile Red LB | ||
| + | #NR, AR, and ATC will spin down tomorrow and resuspended in Nile Red LB | ||
| + | #NR, AR, and ATC | ||
| + | *Add 3ml LB to first 2 tubes | ||
| + | **Then need 25ml LB, add 250ul Arabinose, and 25ul ATC, take out 12ml (leaving 13 reamining), add 6.5ul Nile red and take 12ml into last 4 tubes | ||
| + | |||
| + | |||
| + | |||
| + | ==August 31, 2013== | ||
| + | *Grow up the colonies that Tim transform our biobrick into---have Chloramphenicol resistance so will grow in Chloramphenicol | ||
| + | **Going to try and PCR out our gene---using primers 27 and 30 to make about a 1kb fragment | ||
| + | |||
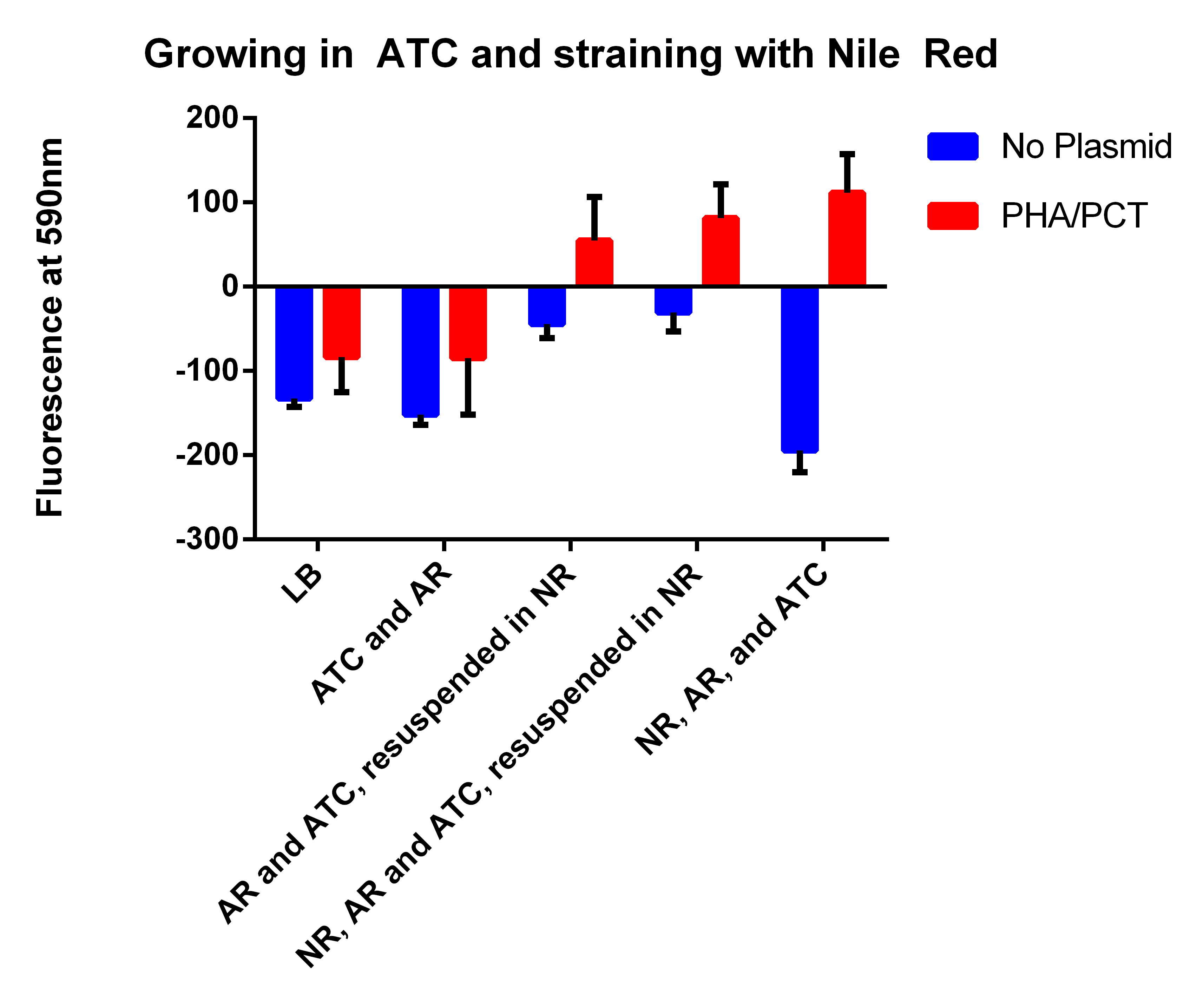
| + | *Going to test letting the cells grow overnight in ATC and Arabinose and staining the next day wth Nile red | ||
| + | **Will test #38 and #1 as a control | ||
| + | #No NR, AR, or ATC | ||
| + | #AR and ATC, will spin down tomorrow and resuspended in LB | ||
| + | #AR and ATC, will spin down tomorrow and resuspend in Nile Red LB | ||
| + | #NR, AR, and ATC will spin down tomorrow and resuspended in Nile Red LB | ||
| + | #NR, AR, and ATC | ||
| + | *Add 3ml LB to first 2 tubes | ||
| + | **Then need 25ml LB, add 250ul Arabinose, and 25ul ATC, take out 12ml (leaving 13 reamining), add 6.5ul Nile red and take 12ml into last 4 tubes | ||
| + | |||
| + | *Also going to repeat ATC and Ethanol experiment | ||
| + | *Need to run an ATC and Nile Red test (to see if it is interfering) | ||
| + | **Going to make 5 test tubes of 1ml Ethanol (80%) | ||
| + | **Adding 2.5ul of Nile red to the 5ml of Ethanol, and placing 1ml in each tube | ||
| + | **Then adding no ATC, 0.5ul ATC, 1.5ul ATC, 3ul ATC, 4.5ul ATC | ||
==August 29, 2013== | ==August 29, 2013== | ||
| Line 604: | Line 782: | ||
[https://docs.google.com/a/yale.edu/spreadsheet/ccc?key=0AmKfFDrNoTUadHlPdkVIMl9fZDlqMTVoUll5TUpJMlE#gid=0 Protocol Here] | [https://docs.google.com/a/yale.edu/spreadsheet/ccc?key=0AmKfFDrNoTUadHlPdkVIMl9fZDlqMTVoUll5TUpJMlE#gid=0 Protocol Here] | ||
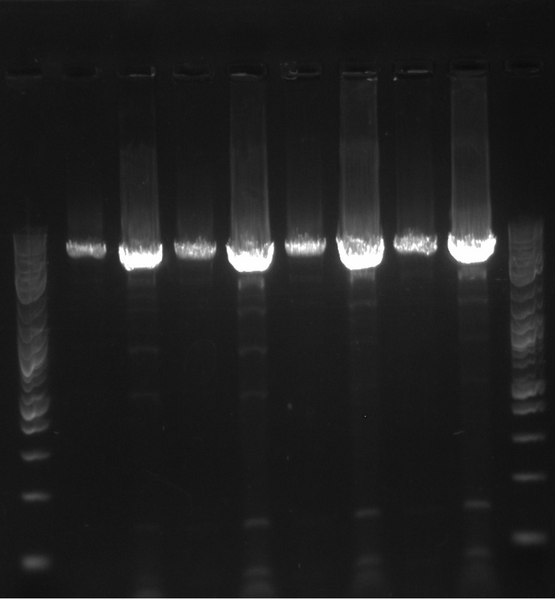
*Let cells grow for 5 hours, and 5/8 showed expected results | *Let cells grow for 5 hours, and 5/8 showed expected results | ||
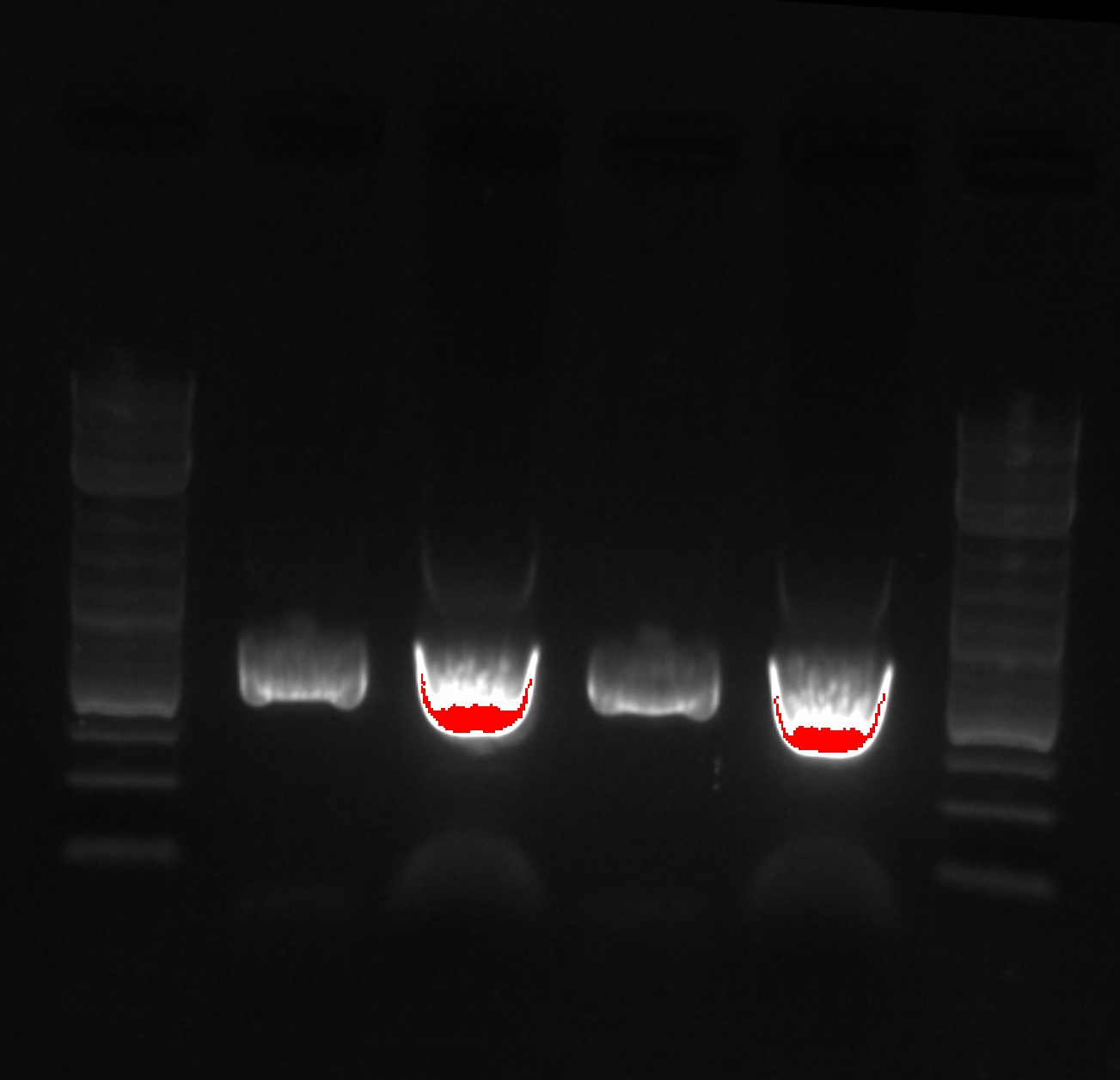
| - | [[File:Capture2.JPG|800px]] | + | [[File:20130909074227!Capture2.JPG|800px]] |
*Plated A1 (ancestor) in 10^-3 and 10^-4 dilutions, Plated A2 (ancestor) in 10^-3 and 10^-4 dilutions, Plated A3 (culture A9) in 10^-4 and 10^-5 dilutions, Plated A5 (A11) in 10^-3 and 10^-4 dilutions, Plated A6 (A11) in 10^-3 and 10^-4 dilutions, | *Plated A1 (ancestor) in 10^-3 and 10^-4 dilutions, Plated A2 (ancestor) in 10^-3 and 10^-4 dilutions, Plated A3 (culture A9) in 10^-4 and 10^-5 dilutions, Plated A5 (A11) in 10^-3 and 10^-4 dilutions, Plated A6 (A11) in 10^-3 and 10^-4 dilutions, | ||
| Line 658: | Line 836: | ||
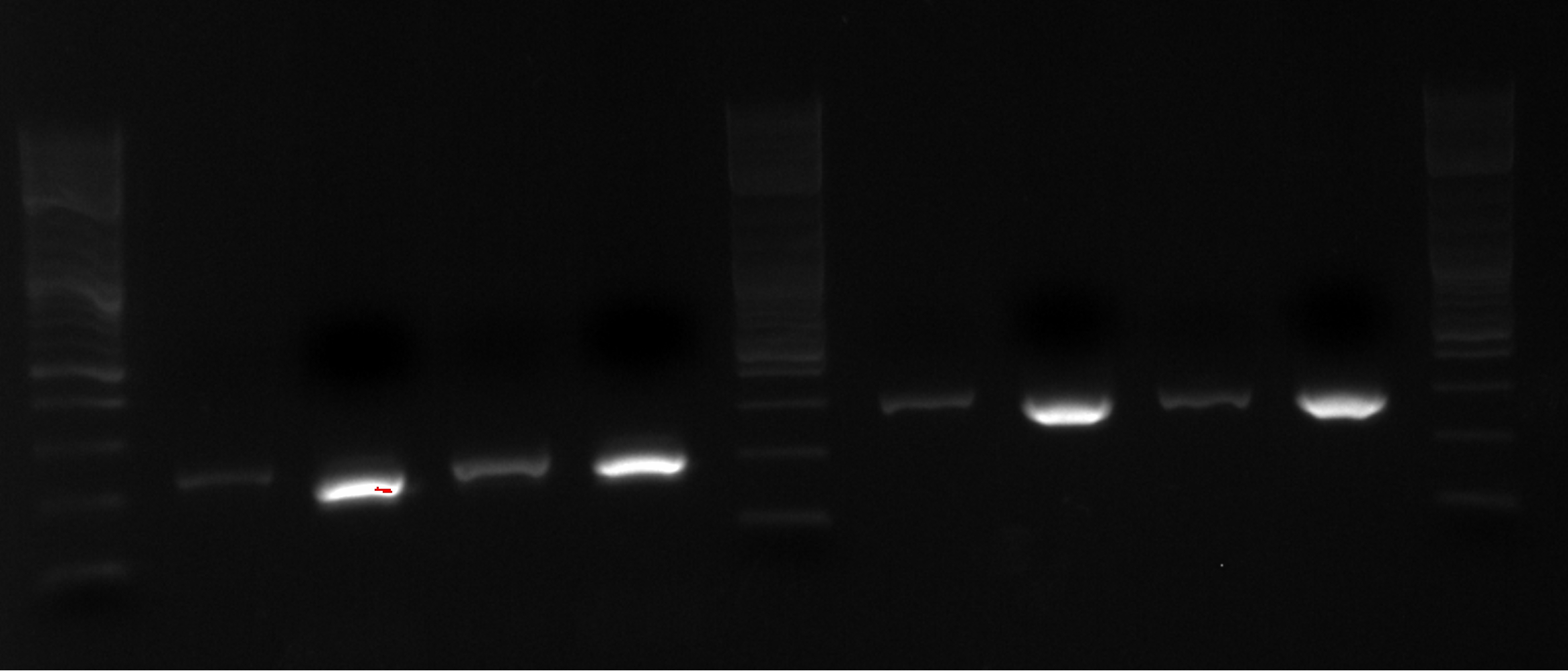
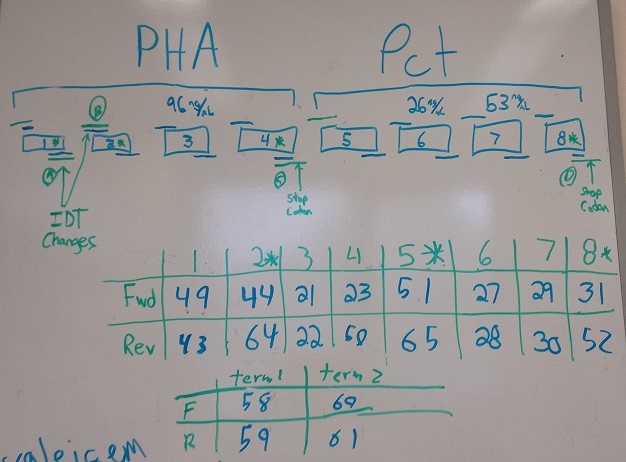
*Designed and ordered primers to put both Biobricks (for export system) into PZE21 plasmid | *Designed and ordered primers to put both Biobricks (for export system) into PZE21 plasmid | ||
*Ordered MAGE oligos | *Ordered MAGE oligos | ||
| + | [[File:PCTTargets.JPG|500px]] | ||
| + | [[File:PHATargets.JPG|500px]] | ||
==July 8, 2013== | ==July 8, 2013== | ||
| Line 1,209: | Line 1,389: | ||
|- | |- | ||
|} | |} | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | '''[[https://2013.igem.org/Team:Yale/Notebook Return to Lab Notebooks]]'''<br> | ||
Latest revision as of 04:42, 16 January 2014
September 27, 2013
- Tested 36, 64, 65, 66 from frozen stocks grown overnight in LB, NR, ATC, Arabinose, Kan
- Spun down 1ml of the cells, washed in PBS once and put 150ul of cells + PBS into 750ul of PBS
- FACS sorted again using PE-Texas red
- Sorted all three tubes according to a gate that had 15 of the baseline (which was 38 or our cells with our plasmid)
- KO had 26, RBS had 65, while All had 98
- We also set a higher gate and sorted the all sample again this time only getting the top 0.1% which was 7/10,000
- I started cultures of all of these and will plate read this weekend
September 20, 2013
- Made new LB and LB agar--for Spec plates later
- Going to test the plLacO clones 68-71, and use 38 as a control, going to put in LB, NR, IPTG, Arabinose, Kan
- 15ml LB, 7.5ul NR, 15ul IPTG, 75ul AR, 15ul Kan
- Start cultures of 64, 65, 66 in LB Kan (Kan is 1000x) for MAGE. These all have our plasmid already
64-->KO oligos, 65---> RBS oligos, 66---all Oligos
- Start 2 cultures of #1 in LB. These don't have our plasmid and would be for later transformations with fixed promoter thing---One is for KO oligos, one for RBS and one for all
September 16, 2013
- My redoing the biobrick did not work, so we just submitted 5.35ul of PCT1 which was at 45.5ng/ul in 10ul of nuc free H2O
- PCT1 is in the plasmid box and in the frozen stock and is a white colony
September 15, 2013
- Running the PCRs of the PCT gene and backbone (2 versions) on a gel--working on making our biobrick
- Gel purified and nanodroped--the DNA tubes are is in the cold block in our -20
- Set up and ran Gibson assemblies
- In PCR 3A on old PCR machines
- Looked at the tetR specR plates and there are 5 or so colonies on the negative control and many more on the positive
- Some sort of contamination present, so will put in the 4 degrees but probably don't want to use
- There is a gibson assembly of plLacO and our construct done---need to transform into EcNR2 and plate on Kan
September 11, 2013
- Setup plate reader control to test all 4 of the positive plLacO constructs
- Growing frozen stock #1, #38, #68-71 in LB NR AR IPTG and as a control growing #38 and 68 in LB NR AR ATC, and 1 in LB NR
- NR is 2000X, ATC is 1000X, Arabinose is 100X, IPTG is 1000X
- Start big culture for PLA extraction
- Going to start a 5ml culture of frozen stock number 38 with 5ul of Kan and let grow overnight
- Also, making a 1L of LB in a 4000ml flask which will be autoclaved
- Then tomorrow need to add the 5ml of confluent cells to the 4L flask and let grow to mid log
- Then add inducers (ATC and arabinose), then let grow overnight
- Redoing PCT amplifications to put on biobrick because we need to do this ASAP
- Going to amplify PCT off of our entire construct (164,165 and 168 and 169)
- Apparently Tim didn't amplify up the biobrick to give it homology to PCT, which we should have done so I just ordered 4 new primers which have the stop codon and amplify up the backbone so the assembly should work a lot better
- bio_PCT_R gggaacctttctcatctctagaagcggccgcgaa
- bio_PCT_F gaaatgaagtccTAAtactagtagcggccgtgcagtccgg
- PCT_Bio_F cggccgcttctagagatgagaaaggttcccattattaccgc
- PCT_Bio_R cggccgctactagtaTTAggacttcatttccttcagacccatt
September 9, 2013
- Here is a graph of the data from switching the promoters
- Does not appear to have worked so probably some of the old plasmid make it through and thus they are really the same---need to DPN1 digest before we transform
- This is the plates from the second attempt at the specR TetR transformation.
- Getting the feeling that these spec plates are bad so going to make new ones and try again
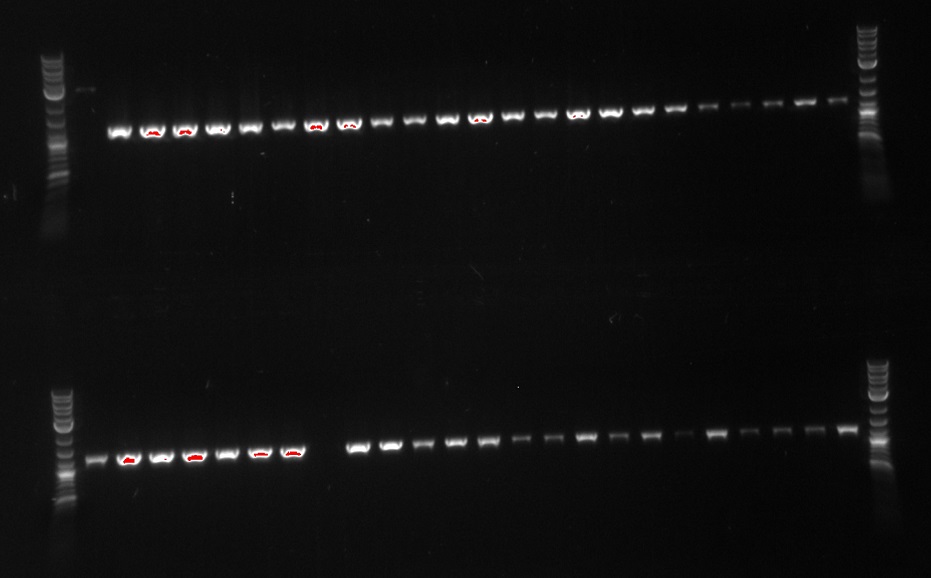
September 7, 2013
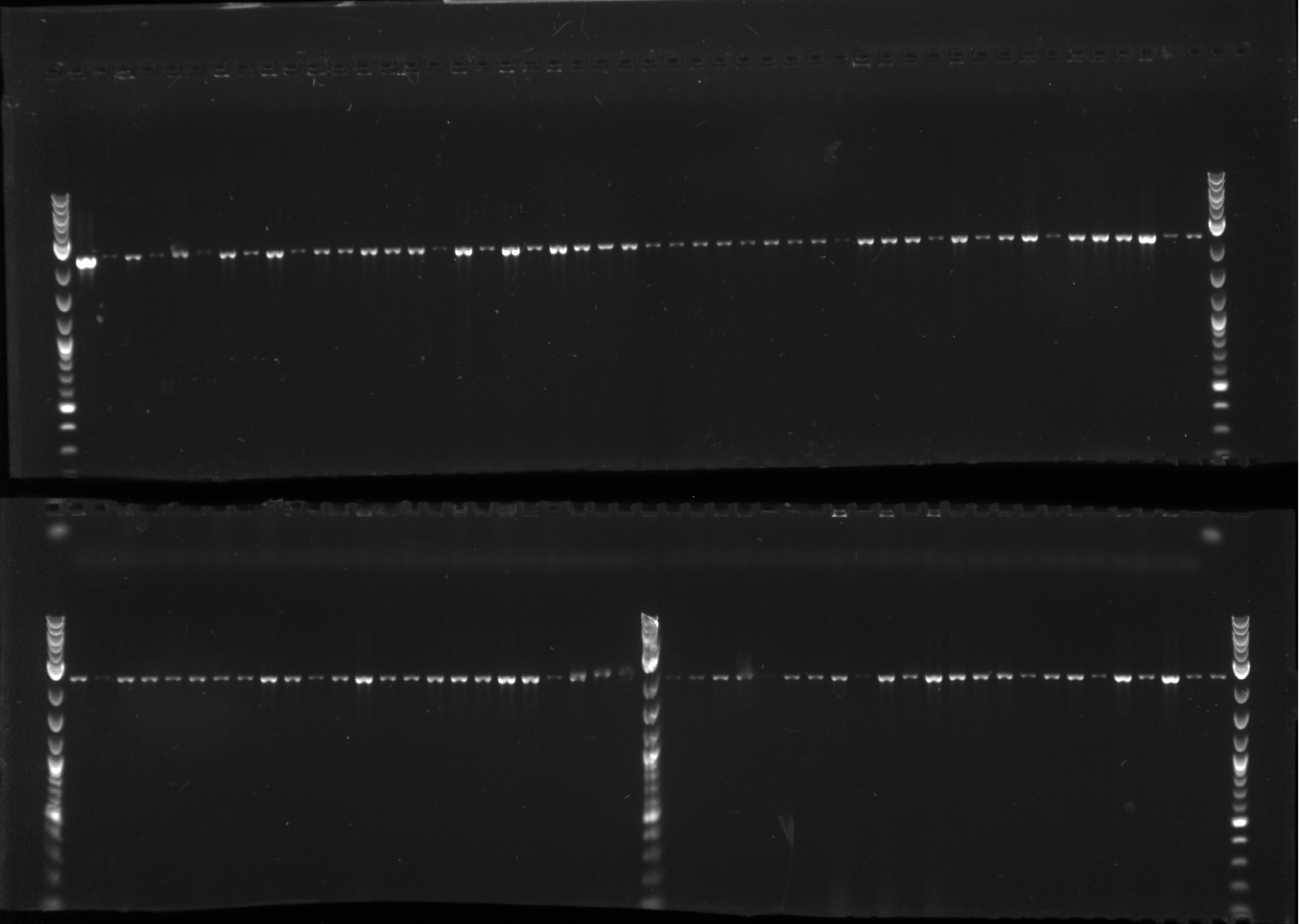
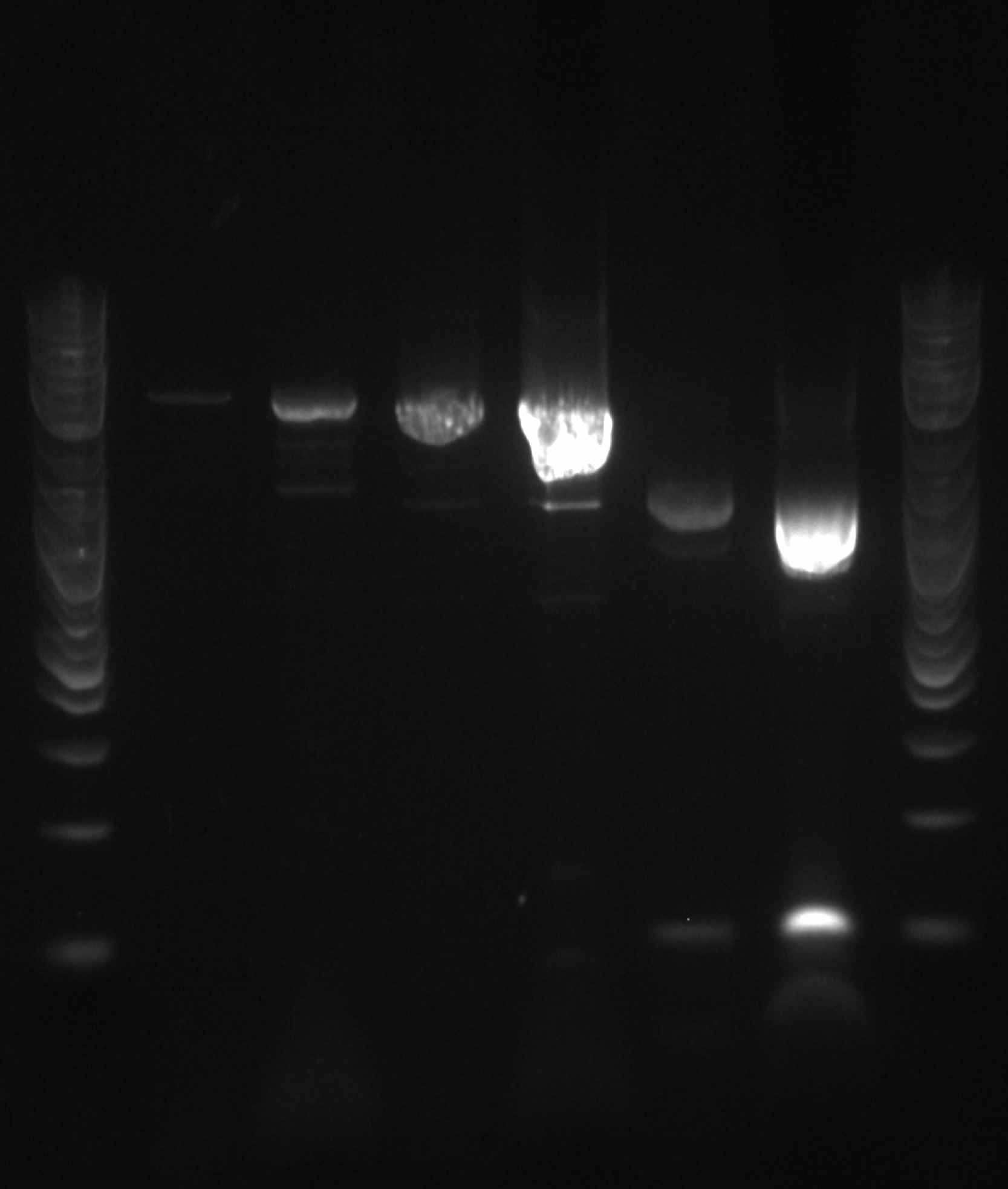
- Picked 46 colonies into 96 well plate---#1 is negative control (EcNR2) and replacing #48 with the DsDNA from Jamin
- primers 172, 173--30 second extension, 65 degrees
- Realized I used the original specR which means homolgoy is only 58 degrees to that so the "positive" control might not work properly
- Run ladder, A1, B1.....ladder in row 1
- Run ladder, C1, D1.....ladder in row 2
- So I grew the Biobricks in Spec by accident and none grew (good sign really since they actually have Cat resistance not spec)
- So regrew all 16 in LB, and will attempt to PCR tomorrow
- Andrew is drop dialyzed and is transforming the gibson assembly from yesterday into EcNR2 #1, and will plate on Kan plates in 1 hour.
September 6, 2013
- Checked spec plates, and there appears to be some colonies, but will let grow overnight to get a clearer picture
- Going to attempt to gibson assemble plLacO promoter and our plasmid minus the plTetO promoter
- Using 3.62ul of the 39.6ng/ul Backbone, with either 1ul or 3ul of the 27.9ng/ul Promoter
- Here are amounts
- Going to transform into EcNR2 #1
- Spun down 50ml of the overnight grown #38 with ATC and arabinose cultures
- Andrew grew in incubator (non-shaking) in a 100ml bottle
- Ben: Bacterial pellets were washed with acetone for 20 minutes
- Growing up all 16 possible cultures which could have PCT on a biobrick---growing them in 1ml LB with 1ul spec
- Will need to PCR test all of them tomorrow--same primers and such Andrew did yesterday
- Also going to need a positive control with the DNA that was used the gibson assembly of the biobrick---try to amplify up the PCT gene with homology to the Biobrick
- Designed and ordered 3 primers to test for spec or TetR
- forward primer: GCATTTATCAGGGTTATGTCCACTGG
- Reverse for tetR: AAGGCGACGTGCGTCCT 1kb
- Reverse for Spec: GGCCAGCAACGTCGGTTC 0.5kb
- Order screening primers so this locus at the genome and look for either spec which is 1.2kb or tetR which is 600bp
- F:GCATTTATCAGGGTTATGTCCACTGG
- R:AGTTTAAATATGGCTGGTTCTCGCAG
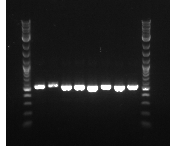
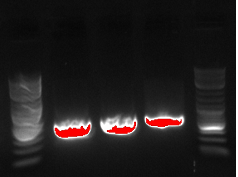
September 3, 2013
- PCR PCT out of Biobrick
- Going to try and PCR out our gene---using primers 27 and 30 to make about a 1kb fragment, 58 degrees, 30 second extension
- PCR 1:PCT1, 2:PCT4, 3:PCT5, 4:PCT8
- PCT1 and PCT 5 were white while PCT 4 and 8 were red
- Redo Spec PCR with Jaymen's plasmid
- PCR 5---primers 172, 173--45 second extension, 58 degress
- Ran a gel: ladder, PCR5, ladder, PCR 1-4 ladder
- Gel purified and nanodroped the SpecR with homology to TetR (22.2ng/ul)---in DsDNA2 box
- Start working on replacing PlTetO with plLacO
- PCR PZE21 6185bp, primers 178,179 58 degrees for 1:45sec extension (6)
- PCR plLacO 136bp, primers 180 and 181, 58 degres for 15 second extension (7)
September 1, 2013
- Going to test Ethanol + NR with different amounts of ATC
- Spun and resuspended cells from yesterday
- Spun down 1ml of the confluent cells, resuspended in LB or LB + NR, and then spun down again and resuspended in LB or LB + NR
- Put back in the incubator for 1 hour (starting at 2pm), and then will test on plate reader
- Will test #38 and #1 as a control
- No NR, AR, or ATC
- AR and ATC, will spin down tomorrow and resuspended in LB
- AR and ATC, will spin down tomorrow and resuspend in Nile Red LB
- NR, AR, and ATC will spin down tomorrow and resuspended in Nile Red LB
- NR, AR, and ATC
- Results
- It appears that the Nile red might have gone bad, so I made new Nile red and used it in these new samples
- Going to test letting the cells grow overnight in ATC and Arabinose and staining the next day wth Nile red
- Will test #38 and #1 as a control
- No NR, AR, or ATC
- AR and ATC, will spin down tomorrow and resuspended in LB
- AR and ATC, will spin down tomorrow and resuspend in Nile Red LB
- NR, AR, and ATC will spin down tomorrow and resuspended in Nile Red LB
- NR, AR, and ATC
- Add 3ml LB to first 2 tubes
- Then need 25ml LB, add 250ul Arabinose, and 25ul ATC, take out 12ml (leaving 13 reamining), add 6.5ul Nile red and take 12ml into last 4 tubes
August 31, 2013
- Grow up the colonies that Tim transform our biobrick into---have Chloramphenicol resistance so will grow in Chloramphenicol
- Going to try and PCR out our gene---using primers 27 and 30 to make about a 1kb fragment
- Going to test letting the cells grow overnight in ATC and Arabinose and staining the next day wth Nile red
- Will test #38 and #1 as a control
- No NR, AR, or ATC
- AR and ATC, will spin down tomorrow and resuspended in LB
- AR and ATC, will spin down tomorrow and resuspend in Nile Red LB
- NR, AR, and ATC will spin down tomorrow and resuspended in Nile Red LB
- NR, AR, and ATC
- Add 3ml LB to first 2 tubes
- Then need 25ml LB, add 250ul Arabinose, and 25ul ATC, take out 12ml (leaving 13 reamining), add 6.5ul Nile red and take 12ml into last 4 tubes
- Also going to repeat ATC and Ethanol experiment
- Need to run an ATC and Nile Red test (to see if it is interfering)
- Going to make 5 test tubes of 1ml Ethanol (80%)
- Adding 2.5ul of Nile red to the 5ml of Ethanol, and placing 1ml in each tube
- Then adding no ATC, 0.5ul ATC, 1.5ul ATC, 3ul ATC, 4.5ul ATC
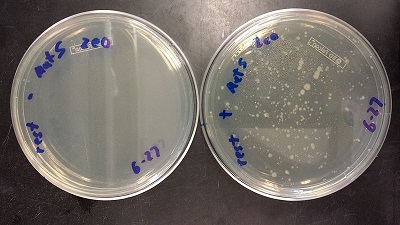
August 29, 2013
- Make Frozen Stock of Spencer's strain
- Run a PCR to amplify Spec with homology to TetR, using primers 172 and 173 and the E. Coli Spencer gave us template DNA
- Spec is 1.2kb so going to run at 58 degrees for 45 second extension time
- No bands appeared on gel

- Going to see if the primers are bad, if not will increase concentration of cells, and remake primers working stocks
- Made Spec plates
August 27, 2013
- Made frozen stock of the MAGE round from yesterday
- Ordered primers to replace plteoO with plLacO and primers to replace tetR with Sep as a selectable marker
August 26, 2013
- Made frozen stocks of MAGE round from Andrew
- Ran another round of MAGE on the cells
- Going to MAGE it with all Oligos, RBS and KO (3 samples)
- Always keeping the cells in Kan, so only those with the plasmid will live
- Left the cells to grow overnight
- Going to MAGE it with all Oligos, RBS and KO (3 samples)
August 8, 2013
- MAGEd cells grow overnight (are confluent), so back diluted and will begin MAGE once they are at mid-log
- Going to MAGE it with all Oligos, RBS and KO (3 samples)
- Always keeping the cells in Kan, so only those with the plasmid will live
- Making a frozen stock after each cycle
- Going to MAGE it with all Oligos, RBS and KO (3 samples)
- Plate read the samples from yesterday
- Take 0.5ml, spinning for 1.5 minutes, resuspend in 1ml PBS, spin for 1.5 minutes, and then resuspend in 1ml, place 200ul in each well
- Raw data here
- Final data here
- With our plasmid is for sure more fluorescent than with (38 vs. 1)
- We were probably right to stop using the MAGE x6 strain
- The KO strain that was FACS sorted actually went down (51 vs. 47), however, the 51 ATC vs. 47 ATC is higher which does make sense because we did sort with ATC in the solution
- Also, tested the Nile Red in Ethanol with different amounts of ATC
- I think it is user error to explain the abnormally high reading at 1.5ul
August 7, 2013
- Going to run MAGE on the FACS sorted KO strain (#51)
- Going to MAGE it with all Oligos, RBS and KO (3 samples)
- Always keeping the cells in Kan, so only those with the plasmid will live
- Running 2 cycles, so these cells will have 3 cycles total as of today (1 before FACS and 2 after)
- The cells are growing really slow (3+ hours and still not at mid-log), also started a culture of 38 if we want to switch
- Going to MAGE it with all Oligos, RBS and KO (3 samples)
- Need to run an ATC and Nile Red test (to see if it is interfering)
- Going to make 5 test tubes of 1ml Ethanol (80%)
- Adding 0.5ul of Nile red
- Then adding no ATC, 0.5ul ATC, 1.5ul ATC, 3ul ATC, 4.5ul ATC
- Need to also test the FACS sorted cells vs. unsorted cells
- Going to run one with and one without ATC
- Need to test FAC sorted KO 1x 51 vs. 47
- Need to test FAC sorted All 6x 50 vs. 46
- Also, need 38 and 1 as controls
- Will make 40ml of liquid
- Adding 20ul of 1mg/ml Nile red to the LB
- 400ul of 20% arabinose
- Those with ATC will get 3ul ATC
August 6, 2013
- Plate read the samples from yesterday, those without Arabinose
- They were all about the same and not really fluorescent
- FACS sorted
- These cells were grown in ATC and Arabinose, next time we should try without ATC because for some reason that is interfering
- Set a gate so that no Baseline cells would be collected, and only those that are better that just the plasmid would be sorted
- Only sorted Sample 3 and sample 6 (sample 4 and 5 didn't have any positives)
- Used PE-Texas Red
- [http://www.icmb.utexas.edu/core/Microscopy/pdf/bd-fluorochrome-info.pdf Here is information about the wavelength]
- Negative is #1 EcNR2 without the plasmid
- Baseline is #38 EcNR2 with our plasmid
- Sample 3 is #46 (MAGE x6 all)
- Sample 4 is #49 (MAGE x1 all)
- Sample 5 is #48 (MAGE x1 RBS)
- Sample 6 is #47 (MAGE x1 KO)
August 5, 2013
- Had weekly lab meeting
- Revealed lots of problems as usual
- We needed to always grow our MAGE strain in Kan otherwise it might lose the plasmid, so the last 6 cycles of MAGE have been in vain
- I back diluted the 6X MAGE culture 6ul in 3ml with Kan, and will make a frozen stock of that but probably will never use it
- Revealed lots of problems as usual
- I started a new culture of #38 from frozen stock in Kan, and will run MAGE on that once it grows up
- Going to MAGE it with all Oligos, RBS and KO
- Used 50ul of the mixtures Andrew made and did 3 didn't oligo pools
- Recovering in Kan LB (finished at 7:11)
- Going to MAGE it with all Oligos, RBS and KO
- Need to grow the final MAGED strain in LB with NR and arrabinose, and then also with ATC
- Growing in LB with 10ul Nile Red, 20ul ATC, 200ul AR
- Growing #1, #38, MAGE x6 all (#46) (6ul), MAGE x1 all (12ul), MAGE x1 RBS (12ul), MAGE x1 KO (12ul)
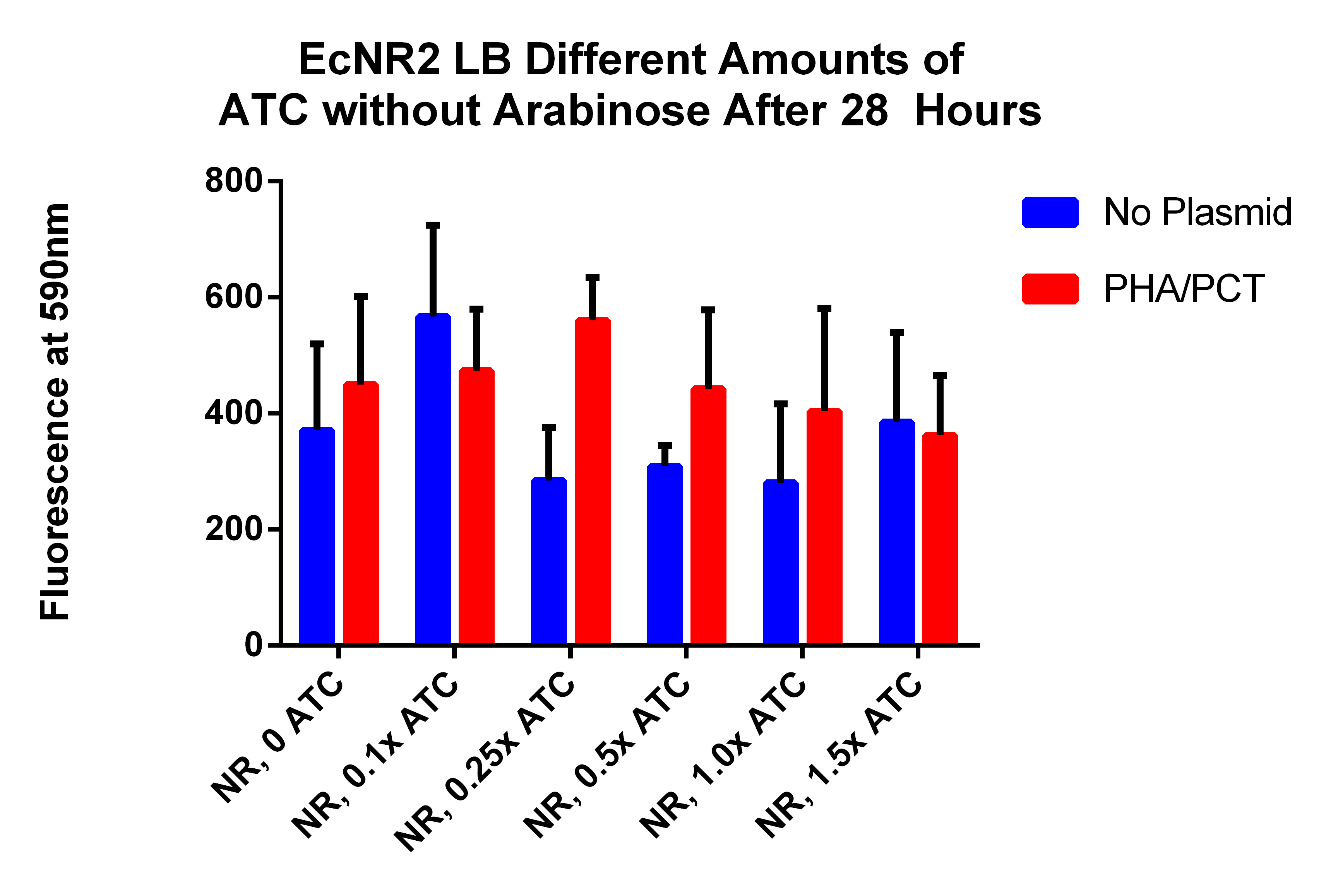
- Going to plate read the 72 hours mark from cultures from friday
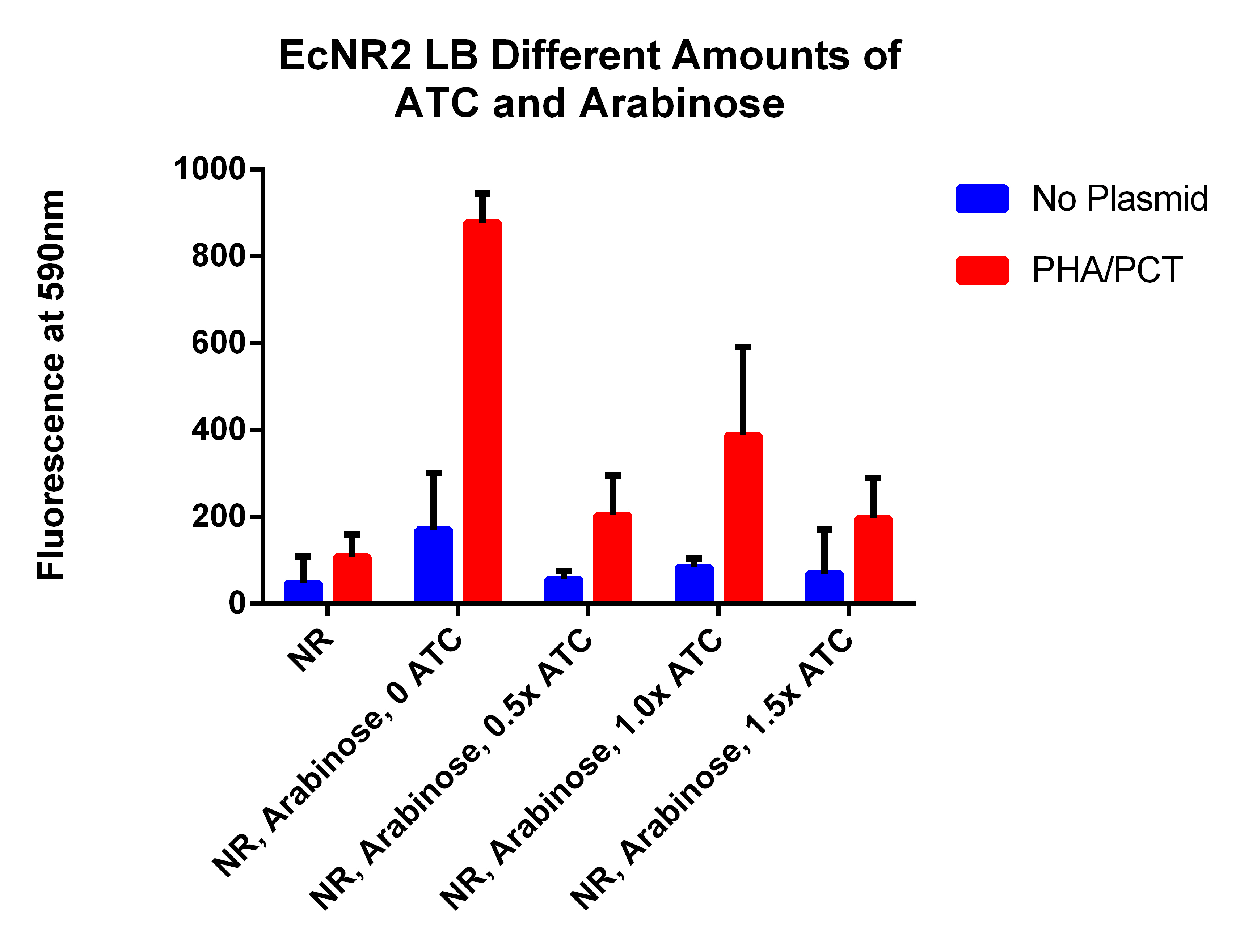
- This is different amounts of ATC to try to understand what is going on (earlier results indicated that adding ATC significantly hurt the fluorescence which is opposite of what we expected)
- Take 0.5ml, spinning for 1.5 minutes, resuspend in 1ml PBS, spin for 1.5 minutes, and then resuspend in 1ml, place 200ul in each well
- This seems like too long of a time and the cells are starting to spazz out, so will not grow cells for 72 hours again
- Data Here
- This is different amounts of ATC to try to understand what is going on (earlier results indicated that adding ATC significantly hurt the fluorescence which is opposite of what we expected)
- Need to make a new plate reader experiment where no arabinose is added, but ATC is in different amounts
- Will make 40ml of liquid
- Adding 20ul of 1mg/ml Nile red to the LB
- Add 3 ml to 12 culture tubes
- 2 will get 0ul of 30ug/ul ATC--AR 0x, ATC 0x
- 2 will get 3 ul of 3ug/ul ATC--AR 0x, ATC 0.1x (1ul ATC in 9ul of water)
- 2 will get 0.75ul of 30ug/ul ATC--AR 0x, ATC 0.25x
- 2 will get 1.5ul of 30ug/ul ATC--AR 0x, ATC 0.5x
- 2 will get 3ul of 30ug/ul ATC--AR 0x, ATC 1x
- 2 will get 4.5ul of 30ug/ul ATC--AR 0x, ATC 1.5x
- Also, going to grow cells is 1X Ar and 1X ATC and add the NR tomorrow
- Two cultures each with 3ml LB, 3ul ATC, and 30ul Arabinose
- Going to grow the cells overnight, then spin down 1ml, resuspended in 1ml LB with Nile Red
- Then let it grow from several hours and then plate read
August 4, 2013
- Transformations plates do not look right, some of the negative have colonies and the positives don't have anything
- Going to place in incubator upstairs till tomorrow morning
- Going to make a frozen stock of the MAGE x5 cells which have been growing on my bench for the last 48 hours, then going to place in incubator till tomorrow because they are surprisingly not that confluent
- Running plate reader at 48 hours, from cultures from friday
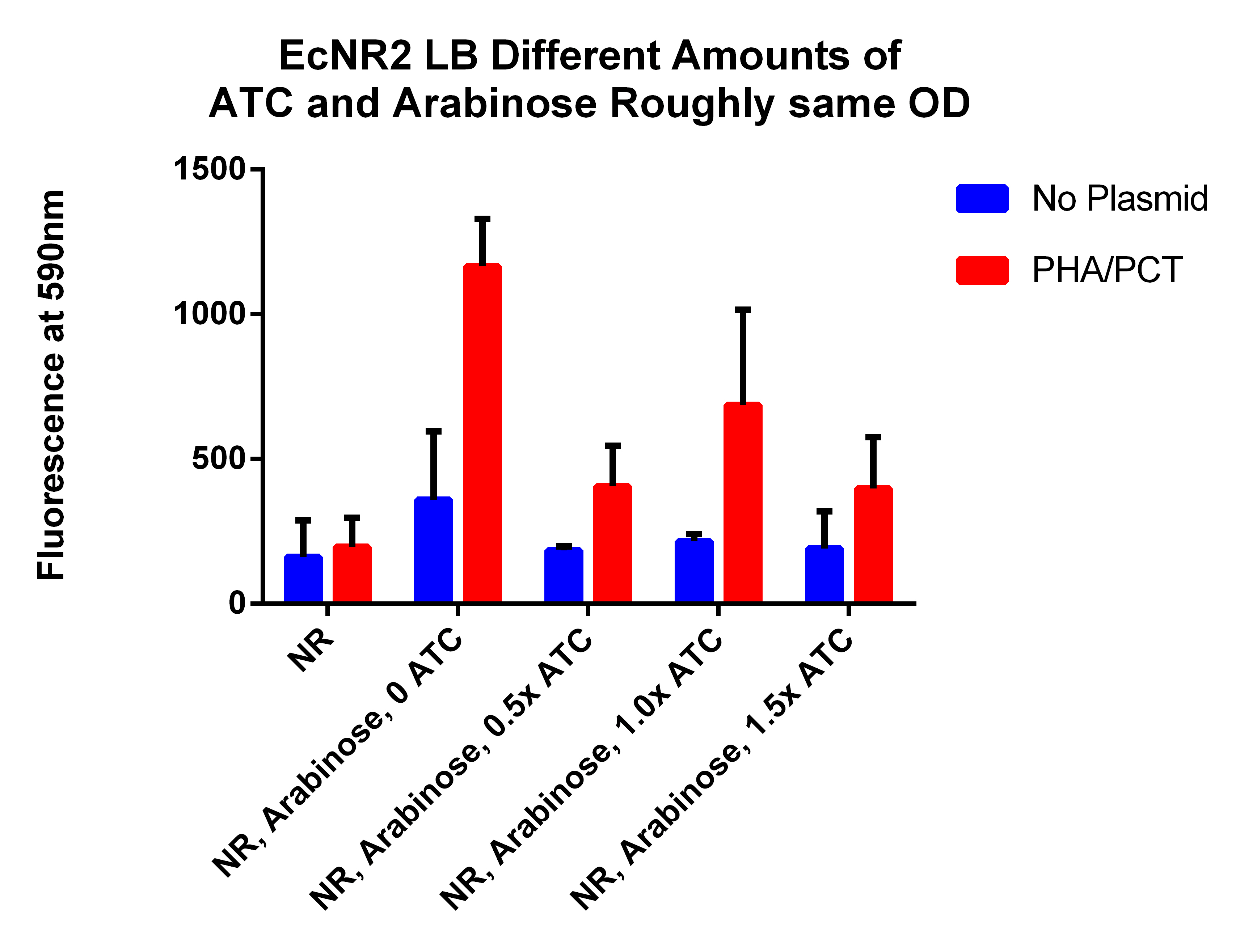
- This is different amounts of ATC to try to understand what is going on (earlier results indicated that adding ATC significantly hurt the fluorescence which is opposite of what we expected)
- Take 0.5ml, spinning for 1.5 minutes, resuspend in 1ml PBS, spin for 1.5 minutes, and then resuspend in 1ml, place 200ul in each well
- Data Here
- This is different amounts of ATC to try to understand what is going on (earlier results indicated that adding ATC significantly hurt the fluorescence which is opposite of what we expected)
- Going to grow 20ml of #38 and #1 in Nile Red and Arabinose
- Added 20ml LB to a 50ml beaker
- Added 200ul 20% Arabinose
- 10ul of Nile Red (1mg/ml)
August 2, 2013
- The transformation left growing overnight does not appear right
- Both the negative and positive have a lawn even though it was plated on Kan
- Tokyo transformation does not appear to have worked because the negative control has colonies, and the positive rest has very few colonies
- The transform from Andrew does appear to work, but honestly do not trust it so I will do the transformation again today
- Going to miniprep the Tokyo bioBrick out of #31 and then transform again into #37 EcNR2 from Natalie
- Got 90ng/ul and 95ng/ul when nanodroped
- Transformed into #37 EcNR2 from Natalie (grown in Carb), used 0.5ul of the 95ng/ul in 50ul of water as DNA
- Going to plate on Chloram plates
- Plate reader positive control
- Take 0.5ml, spinning for 1.5 minutes, resuspend in 1ml PBS, spin for 1.5 minutes, and then resuspend in 1ml, place 200ul in each well
- The ODS were not the same, so added PBS, mixed and removed the same amount of PBS to columns 2,3,7,8
- Going to set up cultures tubes so they can grow until Monday which would be 72 hours
- Will make 46ml of liquid
- Adding 23ul of 1mg/ml Nile red to the LB
- Take out 6ml (3ml into 2 culture tubes)--this is neither gene on
- 400ul of 20% arabinose
- Add 3 ml to 12 culture tubes
- 2 will get 0ul of 30ug/ul ATC--AR 1x, ATC 0x
- 2 will get 3 ul of 3ug/ul ATC--AR 1x, ATC 0.1x (1ul ATC in 9ul of water)
- 2 will get 0.75ul of 30ug/ul ATC--AR 1x, ATC 0.25x
- 2 will get 1.5ul of 30ug/ul ATC--AR 1x, ATC 0.5x
- 2 will get 3ul of 30ug/ul ATC--AR 1x, ATC 1x
- 2 will get 4.5ul of 30ug/ul ATC--AR 1x, ATC 1.5x
August 1, 2013
- Plated the overnight grown recombineered EcNR2 on Kan Plates
- Back dilute these cells---Transform EcNR2 with Tokyo biobrick and plate on Chloramphenicol plates
- Plate reader samples from yesterday
- Taking out 0.5ml from each, spinning for 1.5 minutes, resuspend in 1ml MilliQ water by accident, spin for 1.5 minutes, and then resuspend in 1ml, place 200ul in each well
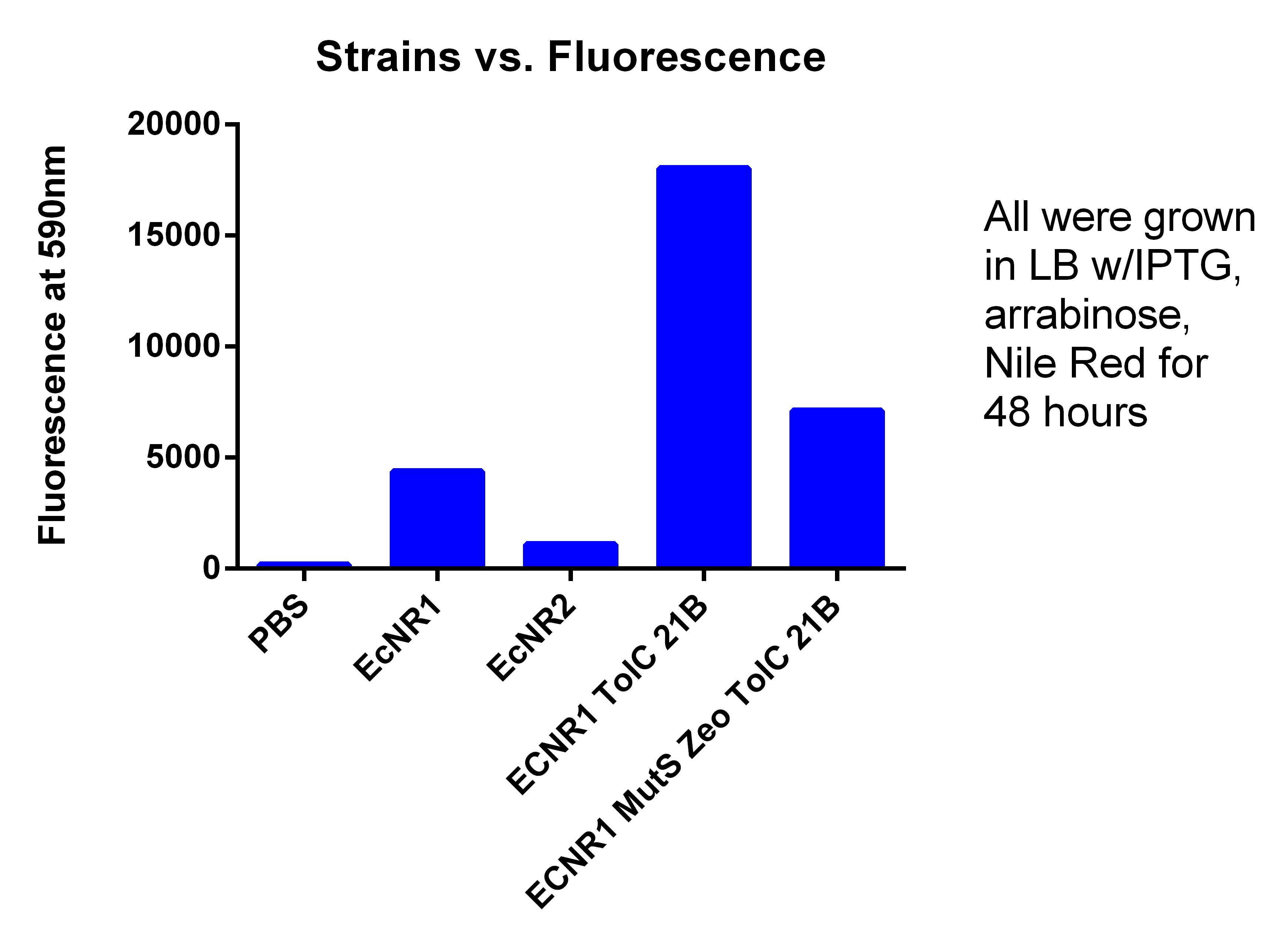
- Results here
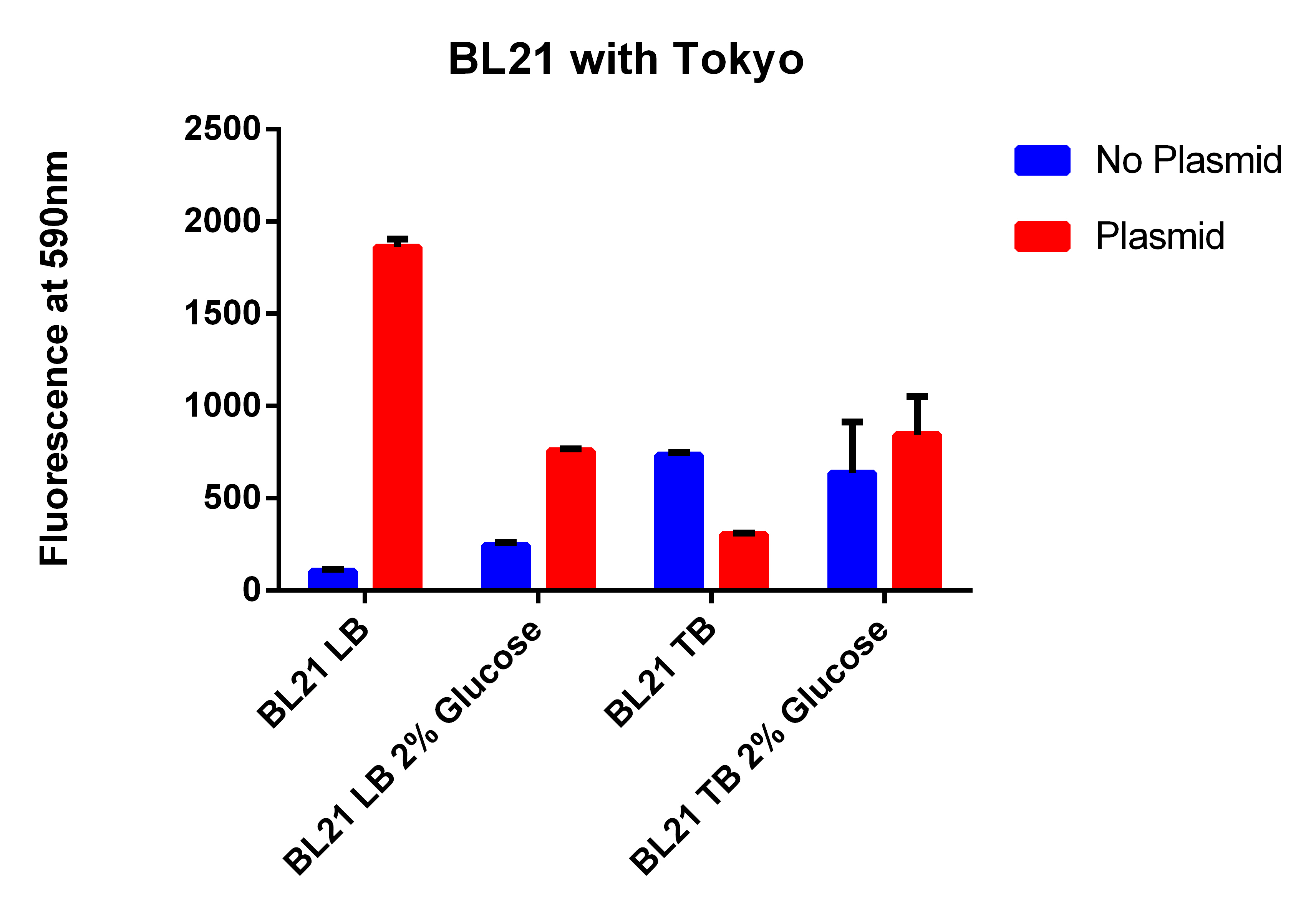
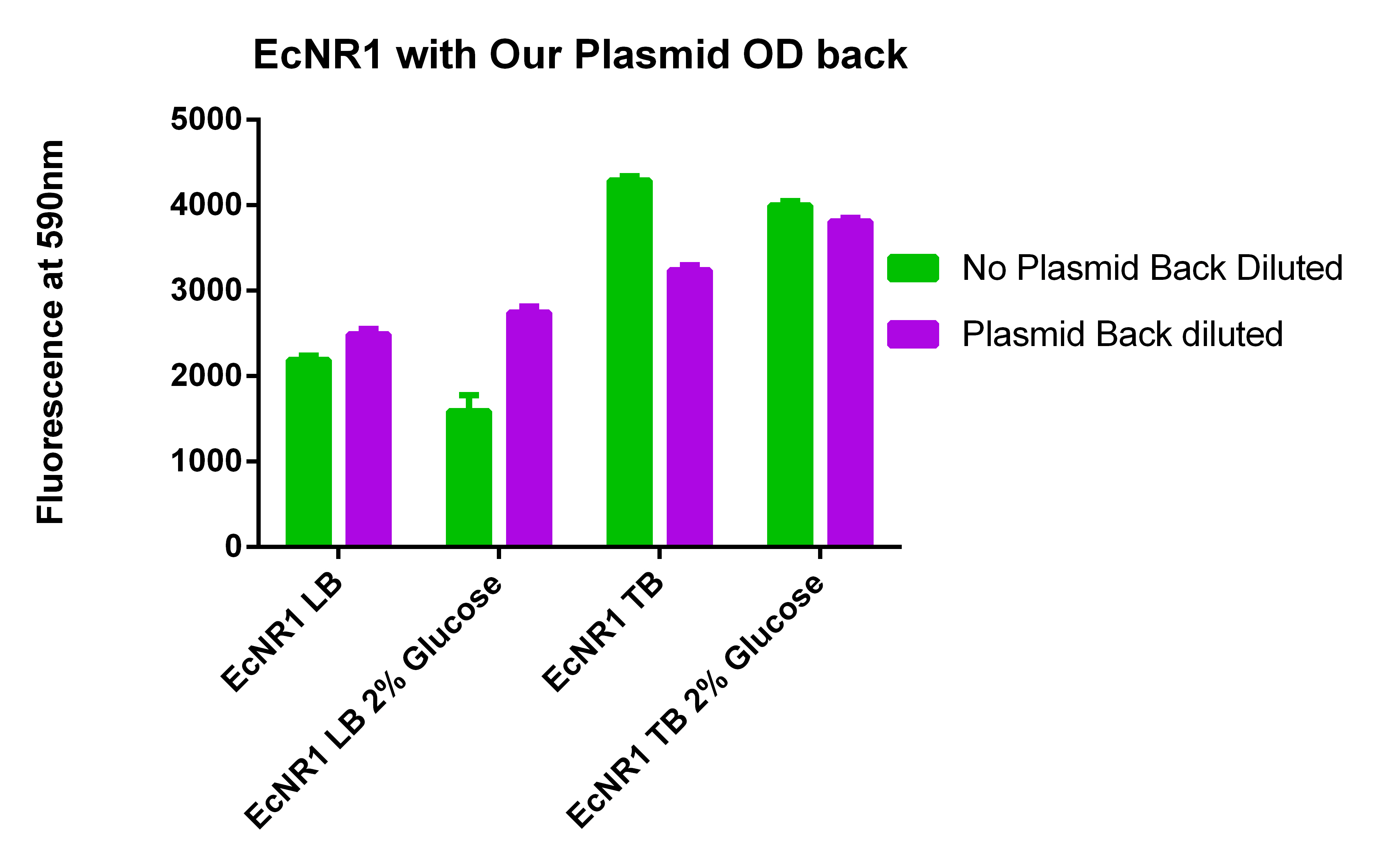
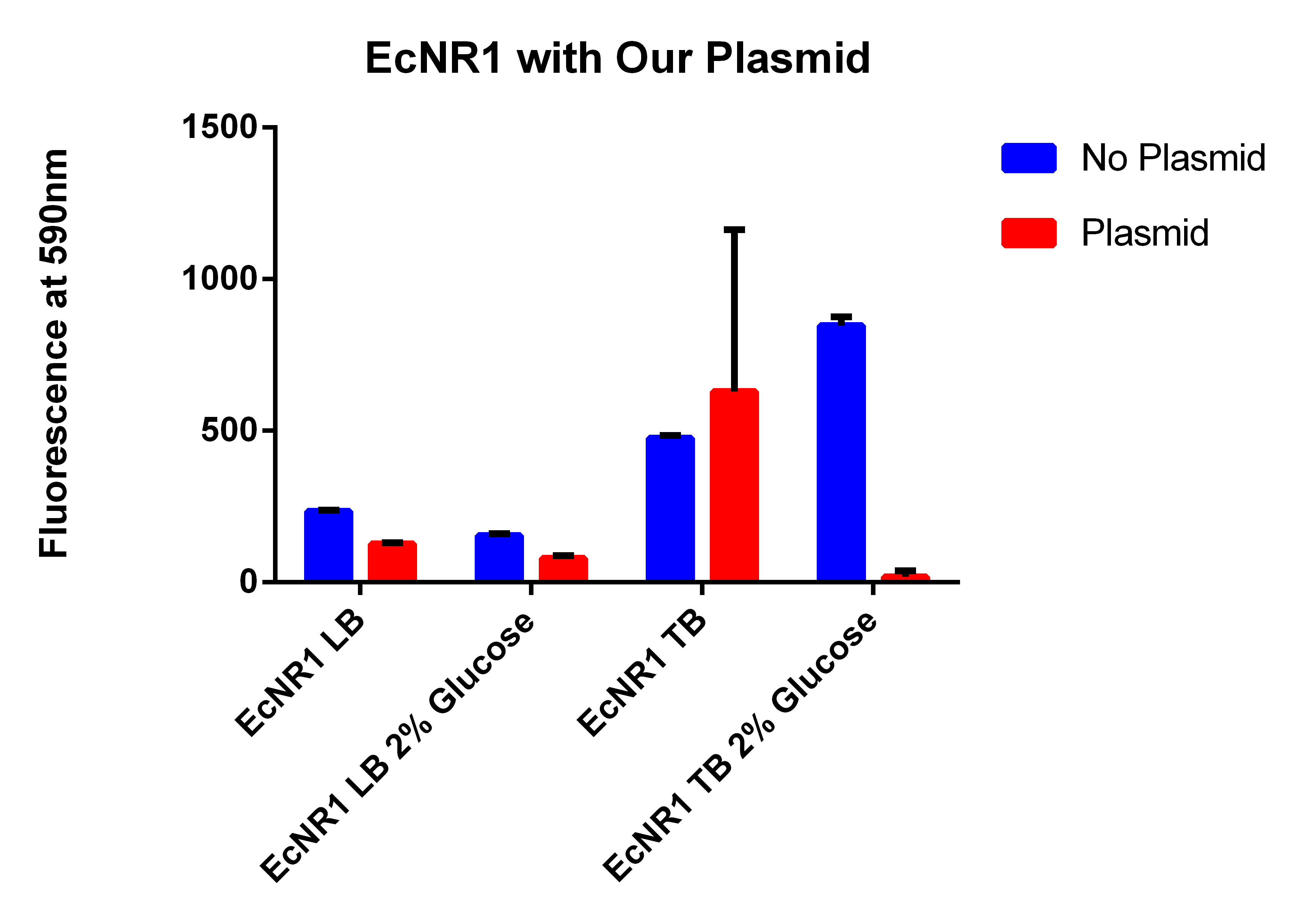
- The BL21 strain with our plasmid does not appear to be working
- The FACS sorted column from today has to be compared with the other EcNR1 MutS Zeo, TolC 21B, so compared it to the data from yesterday, (Therefore hard to compare the data)
- But it does not appear that the FACS sorting helped
- We are not going to use this strain because for some reason its background fluorescence is super high compared to other EcNR2 strains
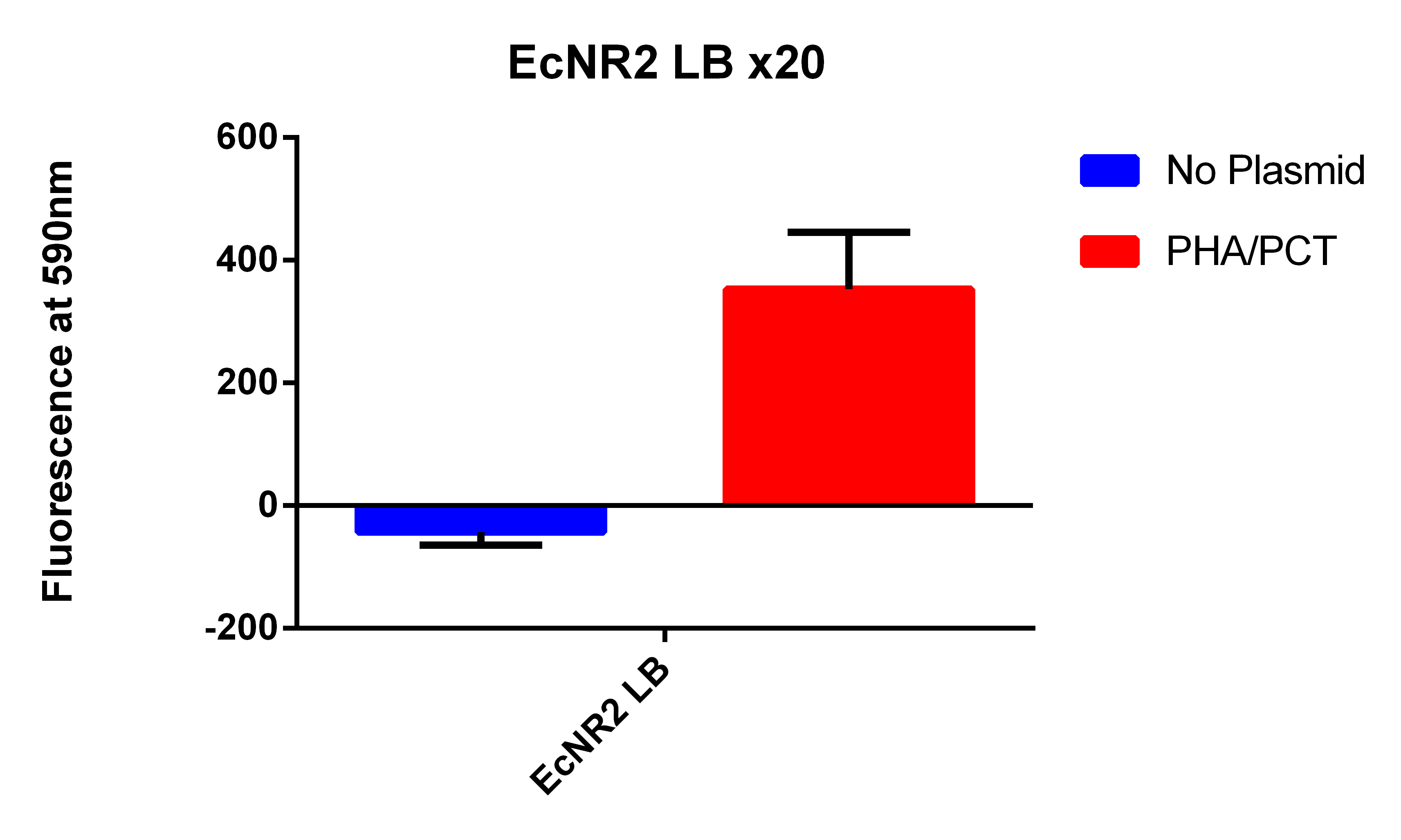
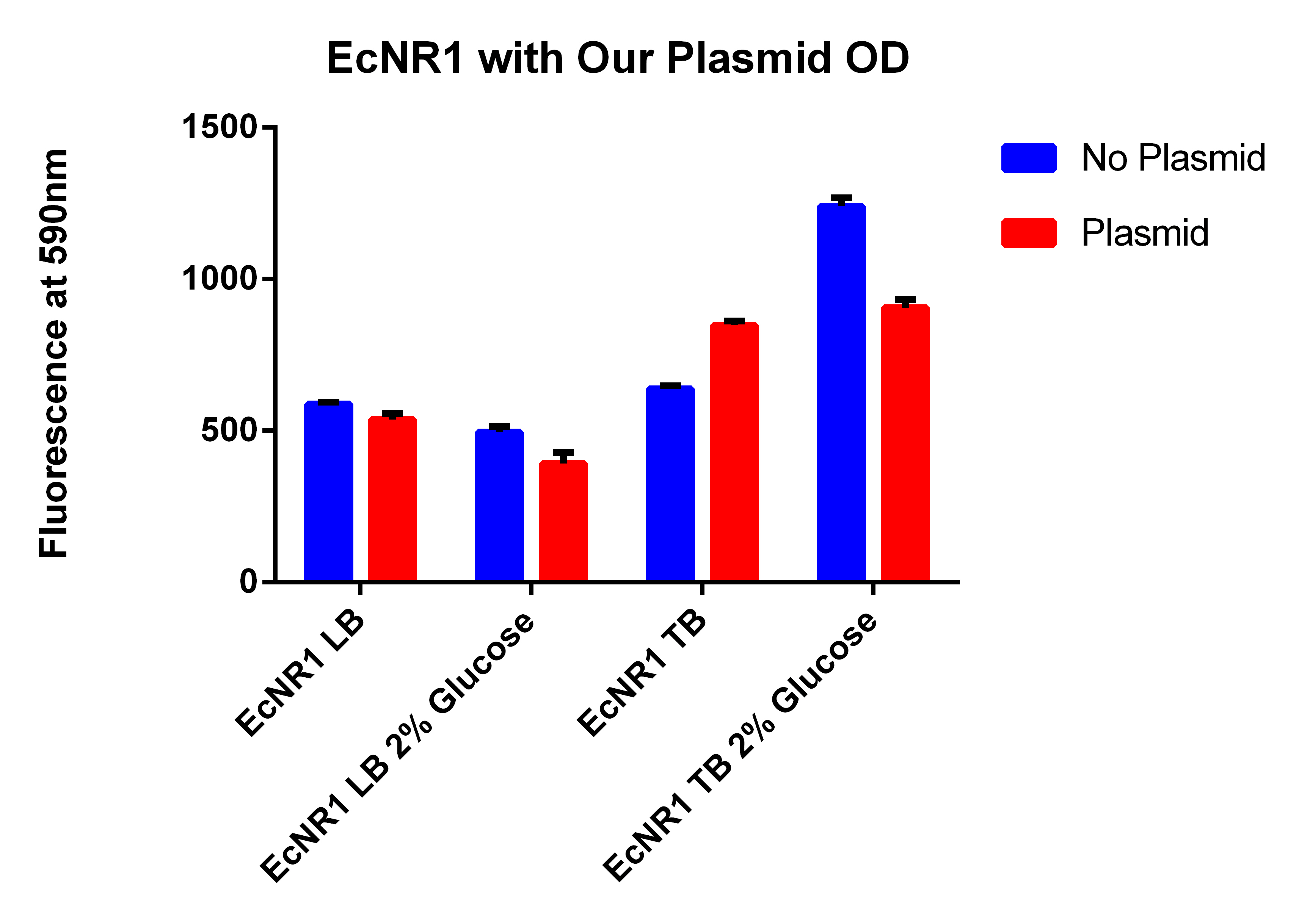
- It looks like EcNR2 with and without our plasmid shows a difference in LB, so taking the remaining liquid in the cultures and repeating the experiment in a large number of samples to make sure the data is significant
- Taking out 0.5ml from without the plasmid and .4ml from the culture with the plasmid to account for the difference in OD
- Spinning for 1.5 minutes, resuspend in 1ml PBS, spin for 1.5 minutes, and then resuspend in 1ml, place 200ul in each well
- Data here
- Taking out 0.5ml from without the plasmid and .4ml from the culture with the plasmid to account for the difference in OD
- OD still were not the same so added 60ul of PBS to the wells with the EcNR2 without plasmid, mixed and then took 60ul out so all well would have the same 200ul
- Even though the one with the plasmid had a lower OD (and thus less cells) it has a higher fluorescence
- Indicates that we are making something that is causing the Nile red to be fluorescence
- Going to need to prove that our Biobrick works aka the PCT gene works
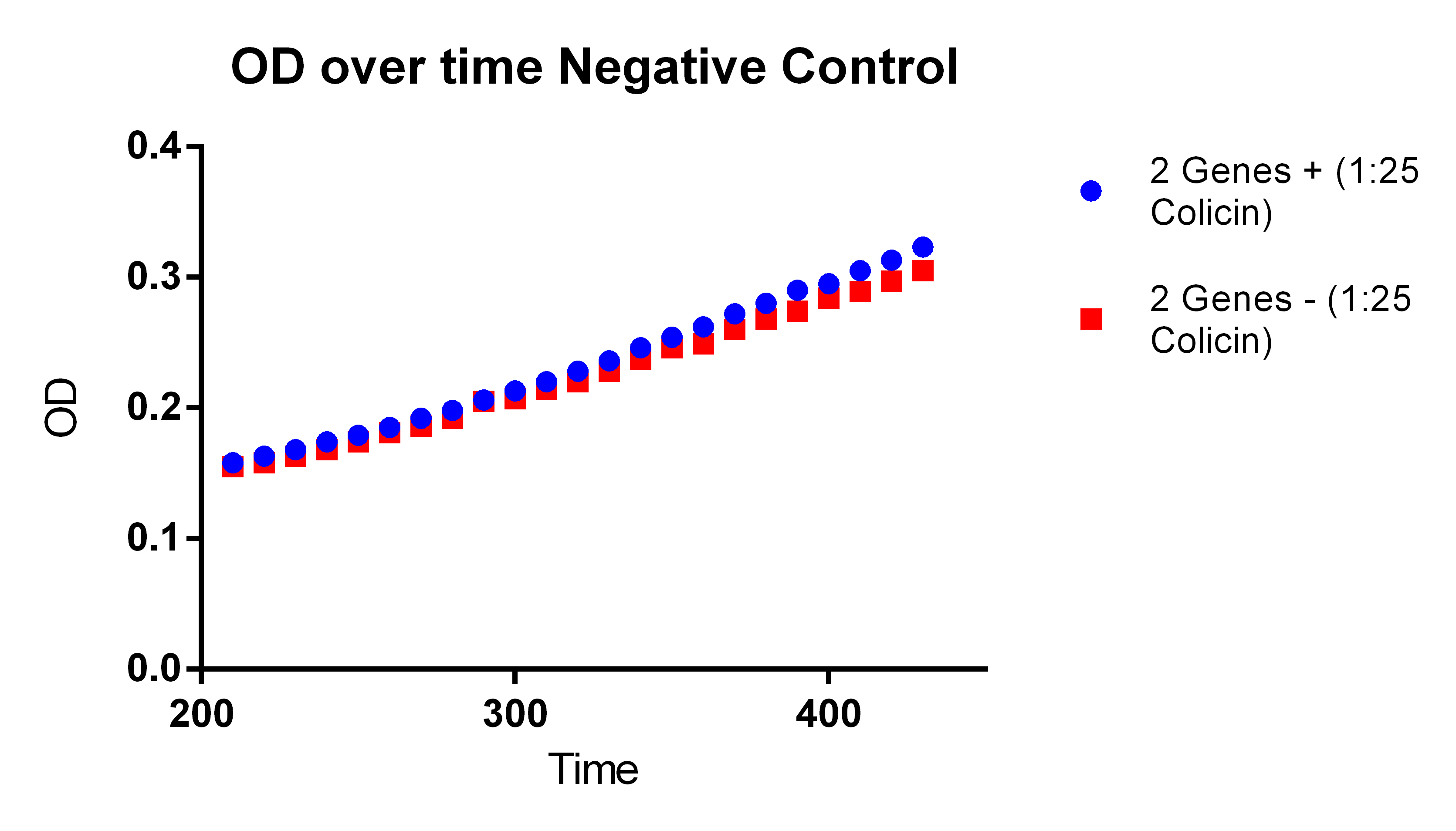
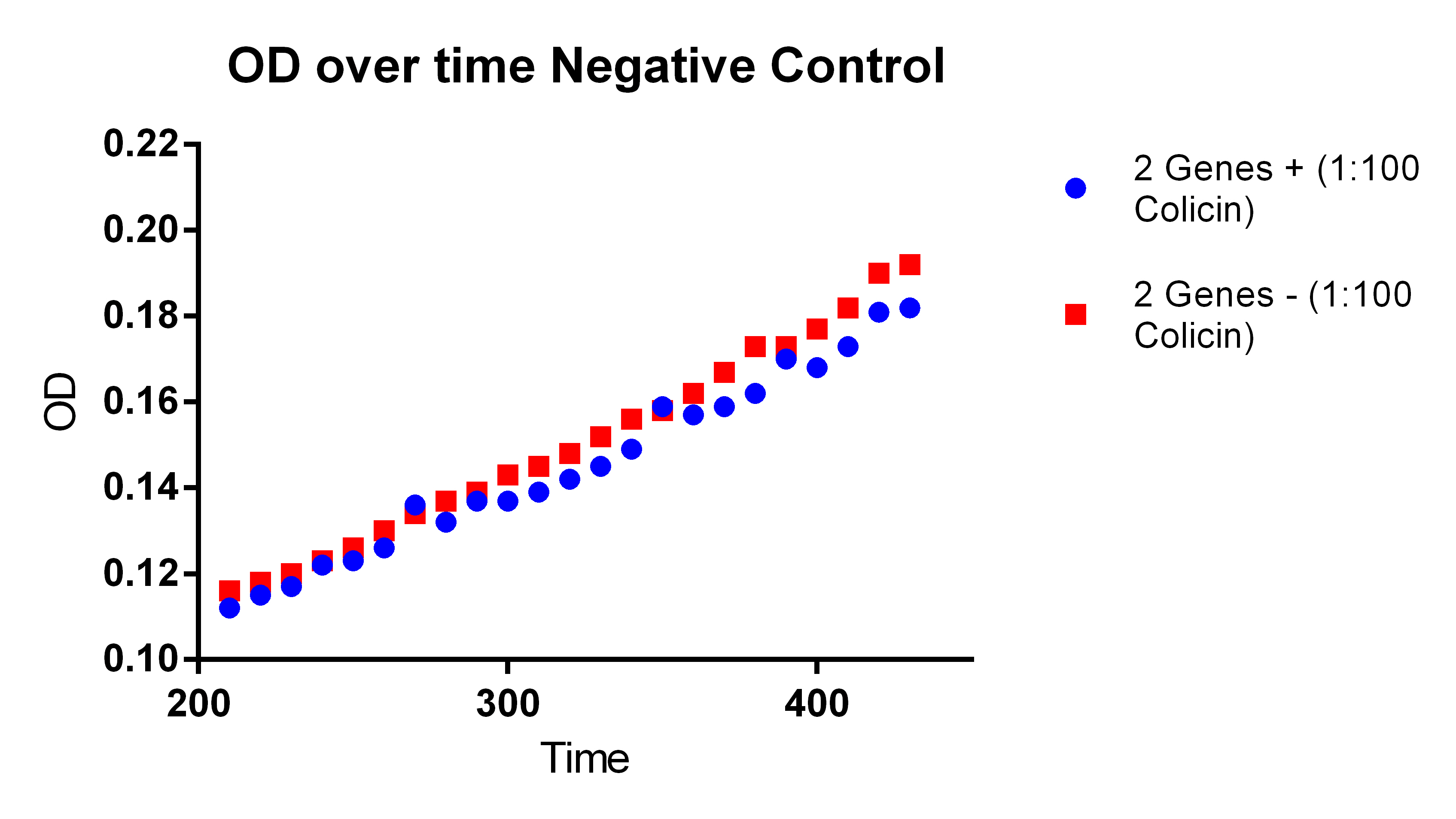
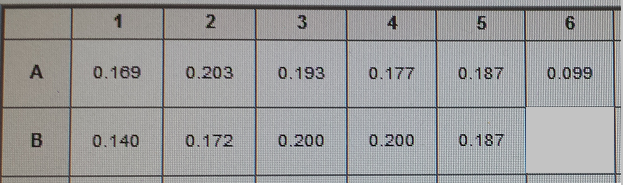
- In order to do this need to experiment where neither genes are on, both genes on (maybe two different amounts of ATC to control the PCT), and then only PHA on
- This will show that the PCT increases the fluorescence and thus the PLA and tada it works
- In order to do this need to experiment where neither genes are on, both genes on (maybe two different amounts of ATC to control the PCT), and then only PHA on
- Making 4 cultures for EcNR2 with our plasmid (#38) and without our plasmid (#1)
- Will make 40ml of liquid
- Adding 20ul of 1mg/ml Nile red to the LB
- Take out 6ml (3ml into 2 culture tubes)--this is neither gene on
- 360ul of 20% arrabinose
- Add 3 ml to 8 culture tubes
- 2 of the 8 will get 3ul of 30ug/ul ATC--this is both genes on
- 2 of the 8 will get 1.5ul of 30ug/ul ATC--this is both on but PCT on lower
- 2 of the 8 will get 4.5ul of 30ug/ul ATC--this is both on but PCT on higher
- 2 of the 8 will get 0ul of 30ug/ul ATC--this is only PHA on
July 31, 2013
- Tokyo group used 20mM Pantothenic acid
- To make a 10X solution that would be .2M (or 2.3827g in 50ml)
- Use plate reader to read the fluorescence of the samples from yesterday
- Data is here
- FACS sorted a sample of #36 (EcNR1 MutS Zeo with our plasmid)
- Found that PE or Phycoerythrin was the best detector which is ex. 495 and em. 545
- Here are the pdf's of the data
- We sorted according to the gates shown and are growing a culture of the sorted FACS results
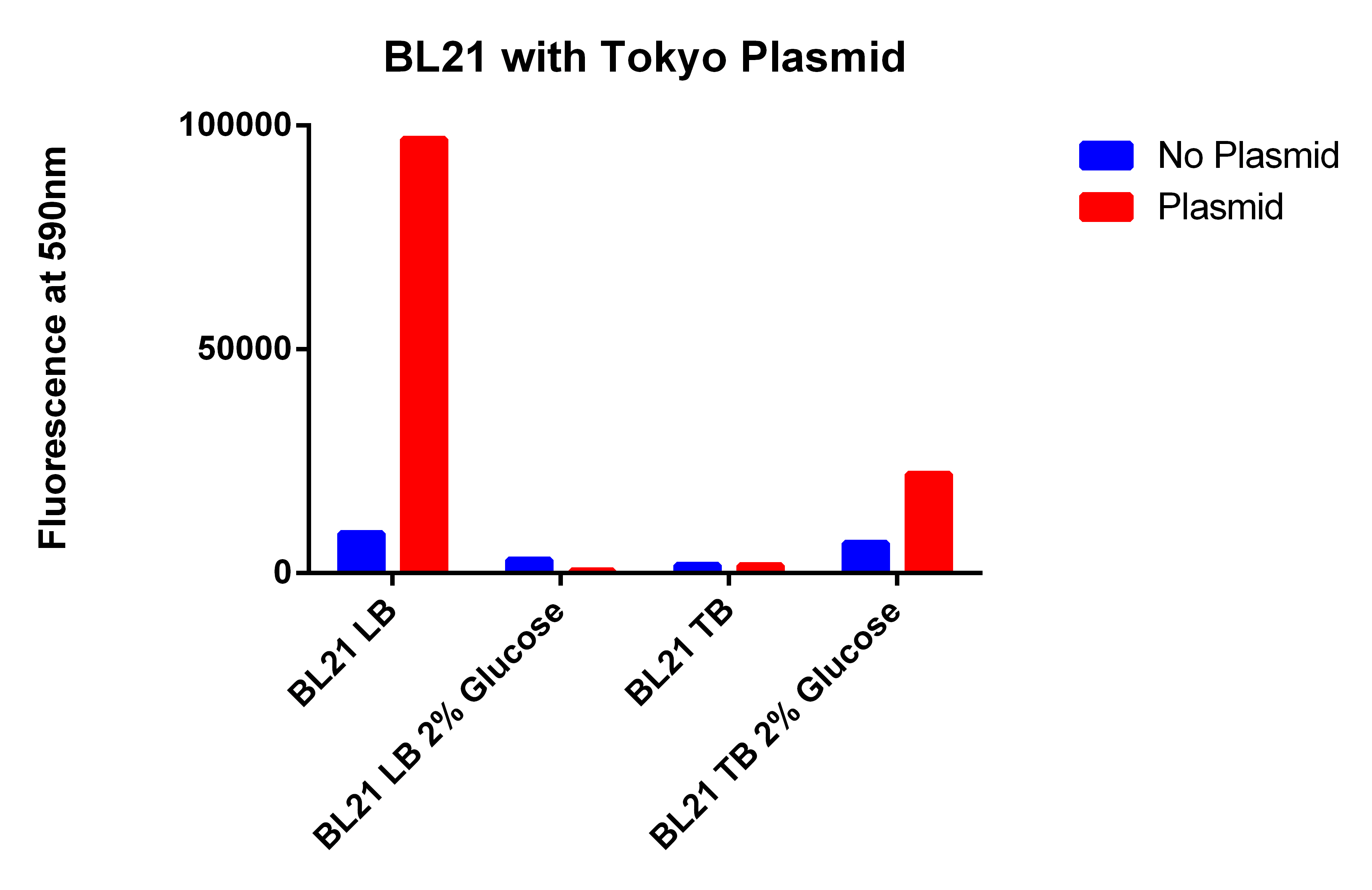
- Need to make cultures with EcNR2 with (#38) and without (#1) our plasmid, and FACS sorter plasmid (#40)
- Need to make cultures of BL21 (#39) with and without (#5) our plasmid, along with the Tokyo BioBrick (#31)
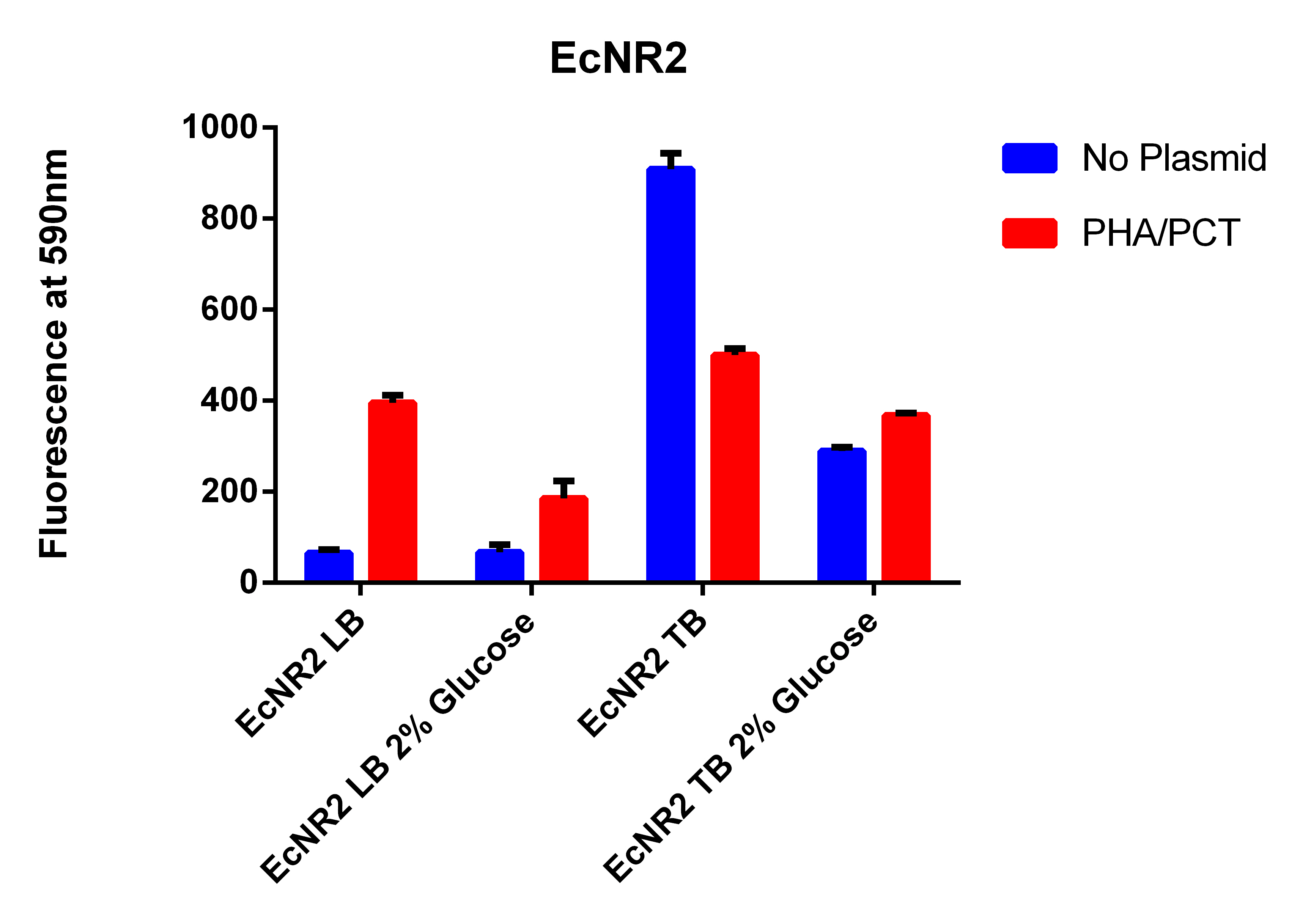
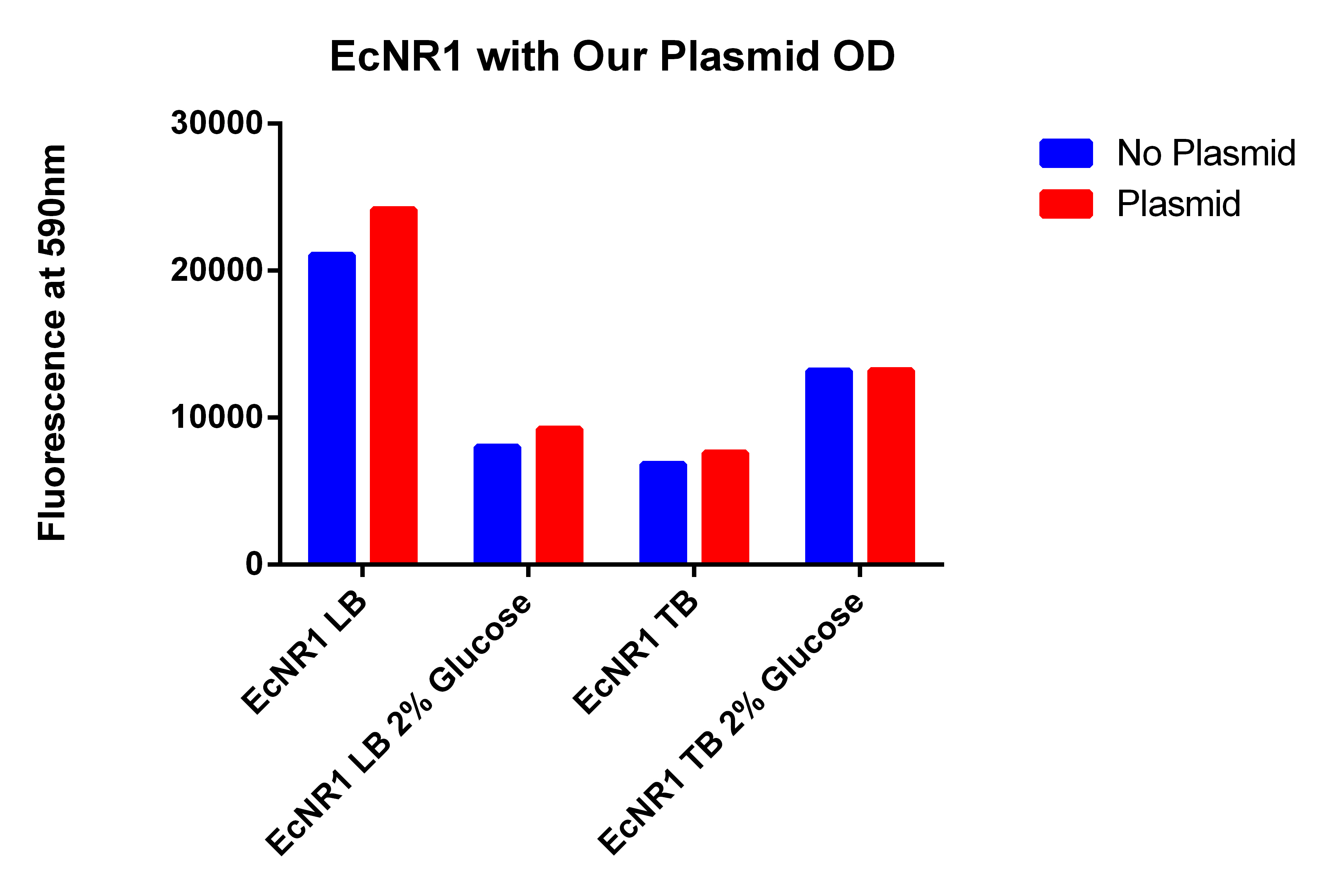
- Going to grow all 6 strains in LB and TB with and without glucose
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Some will get 300ul of 20% in a 3ml Culture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Making small batches of TB mixture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Some will get 300ul of 20% in a 3ml Culture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Going to grow all 6 strains in LB and TB with and without glucose
- Transforming the Tokyo Biobrick didn't work (probably from the bad miniprep)
- Transforming our construct into EcNR2 and BL21 worked
- Andrew is adding to frozen stock
- And Starting to MAGE the EcNR2 with out Biobrick with the RBS and KO oligos
- Found out that can't use our full construct for the Biobrick because it can't have any restriction sites
- Luckily the second gene alone (PCT) doesn't have any sites so ordered primers to put that on a BioBrick
- Recombineering EcNR2 with out construct plus KanR
- Using 3ul of 66ng/ul DNA in 50ul Water
- Going to let one recover for a couple hours and the other recover over night to hopefully increase yield
- Finished at 5:20pm
July 30, 2013
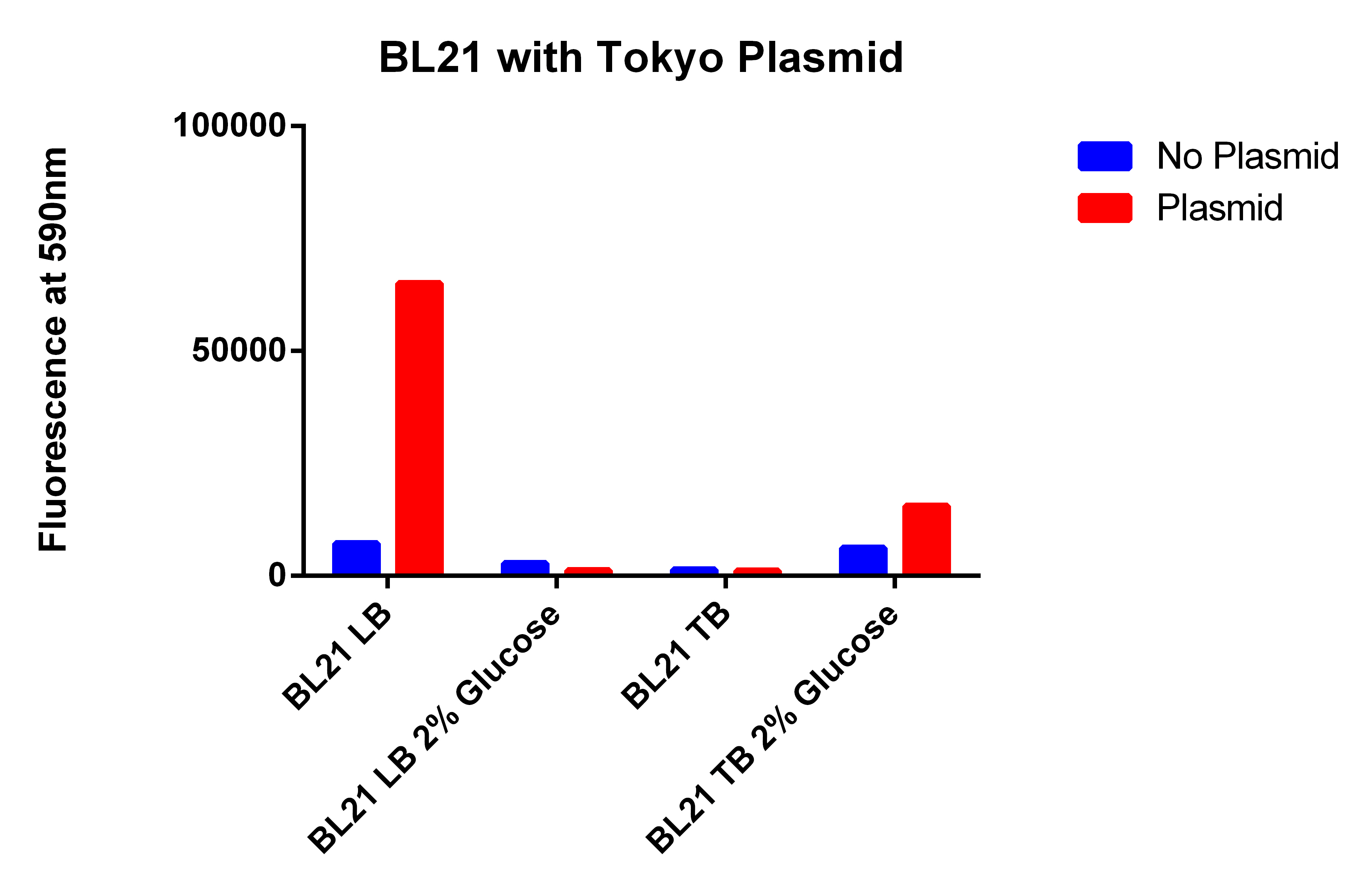
- Use plate reader to read the fluorescence of the samples from yesterday
- LB glucose cells did not grow nearly as quickly as the others (very hard to get a pellet)
- Spun for 2.5 minutes, then 1.5 and 1.5 minutes
- Took 0.5ml of cells and resuspended in 1ml and placed 200ul in each well
- Something clearly did not work properly because the fluorescences from before were 15,000 range for even the normal
- I think the Nile red might have gone bad
- Repeating experiment from yesterday because results this morning were weird and don't look right
- Using EcNR1 MutS Zeo TolC 21B, and EcNR1 MutS Zeo TolC 21B w/our construct ad BL21 and BL21 with Tokyo Biobrick
- In LB, Nile Red, ATC and Arabinose
- Making small batches of LB mixture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Some will get 300ul of 20% in a 3ml Culture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Making small batches of TB mixture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Some will get 300ul of 20% in a 3ml Culture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Taking from frozen stocks #36 and #11 for ECNR1
- Taking from frozen stocks #31 and #5 for ECNR1
- Also, back diluting the 8 samples from today 1:100 to see if the extra time produces more PLA
- Andrew did the transformations
- Transform Bl21 with our construct (3ul of the Gibson in 50ul water) on PZE21 (grow on Kan plates)
- Transform EcNR2 with our construct (3ul of the Gibson in 50ul water) on PZE21 (grow on Kan plates)
- Transform the EcNR2 from Natalie with the Tokyo Biobrick (36ng of DNA) and (grow on Chloramphenicol plates)
- Ordered new primers to amplify up our construct to put on Biobrick
- This is only our genes without KanR, and also ordered primers to amplify the backbone with homology to our construct as a back up plan
- Going to amplify our construct with KanR at the end with the primers that come in today
- Use primers 152 and 153 HiFi and 2:45 extension time at 60 degrees and looking for a 5kb fragment
- Running 1 with no DMSO, 1 with .5 ul DMSO (2%), 1 with 1.25 ul DMSO (5%)
July 29, 2013
- Need to use ATC not IPTG, sadness
- Repeating experiment I set up earlier with both EcNR1 MutS Zeo TolC 21B, and EcNR1 MutS Zeo TolC 21B w/our construct on plasmid in LB, Nile Red, ATC and Arabinose
- Making small batches of LB mixture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Some will get 300ul of 20% in a 3ml Culture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Making small batches of TB mixture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Some will get 300ul of 20% in a 3ml Culture
- Adding 20ul of 1mg/ml Nile red, 400ul of 20% arrabinose, and 40ul of 30ug/ul ATC
- Taking from frozen stocks #36 and #11
- Making small batches of LB mixture
- Growing BL21, EcNR2 and EcNR2 (From Natalie with Carb resistance)
- Going to transform Bl21 with our construct on PZE21 (grow on Kan plates)
- Going to transform EcNR2 with our construct on PZE21 (grow on Kan plates)
- Going to transform the EcNR2 from Natalie with the Tokyo Biobrick and (grow on Chloramphenicol plates)
- Running a PCR to amplify our construct (PHA and PCT) to give it the biobrick prefix and suffix
- Using primers 148 and 149 and going to use HiFi and 2:30 extension time at 58 degrees and looking for a 4.2kb fragment
- Will run in duplicate to increase yield
- The strains we plated yesterday (EcNR1 on TB glucose) didn't fluorescence under the blue light
- The Tokyo did when Andrew did it so we are either not producing PLA or producing it in low quantities
- Since it appears that our strain with our plasmid showed an increase fluorescence vs. the strain without our plasmid repeating this experiment
- Started 3 2ml cultures for the EcNR1 MutS Zeo TolC 21B, and EcNR1 MutS Zeo TolC 21B w/our construct on plasmid
- Will use plate reader and see if the ones with our plasmid are still more fluorescence when run in larger sample size
- Started 3 2ml cultures for the EcNR1 MutS Zeo TolC 21B, and EcNR1 MutS Zeo TolC 21B w/our construct on plasmid
- Designed and ordered primers to sequence the KanR at the end of our two genes, also designed primers to put both of our genes + KanR on a BioBrick
July 28, 2013
- All 8 of Tims colonies from the Kan plates after recombination with out gene+KanR were able to grow in SDS, which means they all have functioning TolC
- Plated each of the 7 colonies plus a control on Kan plates
- All of the colonies were able to grow and control was not
- I probably labeled them wrong, because pretty sure that the control cannot grow and as labeled it did grow and 7 did grow which doesn't make any sense
- Took data from EcNR1, EcNR2, EcNR1 TolC 21B, EcNR1 MutS Zeo TolC 21B all were growing in LB w/Nile red, IPTG, Arabinose
- Took data from EcNR1 MutS Zeo TolC 21B, EcNR1 MutS Zeo TolC 21B w/our construct on plasmid, BL21, and BL21 with Tokyo plasmid
- Each the 4 strains in LB and TB w/Nile red, IPTG, Arabinose with and without glucose
- Procedure, take 1ml of confluent cells, and spun down, washed two times with PBS, and resuspended in 600ul PBS, placed 200ul of PBS
July 26, 2013
- Took 100ml from both TB, Nile red, IPTG, Arabinoseand LB, Nile red, IPTG, Arabinose into a new container and added 10ml of the 20% glucose
- Both the Korean and the Tokyo group supplement with glucose so we need to to produce PLA
- Set up positive control
- Growing EcNR1 MutS Zeo TolC 21B, EcNR1 MutS Zeo TolC 21B w/our construct on plasmid, BL21, and BL21 with Tokyo plasmid
- Growing each of the 4 strains in LB and TB w/Nile red, IPTG, Arabinose with and without glucose
- Diluting 1ul of the cells into 3ml of broth so they have plenty of time to grow--In incubator around 11am Friday
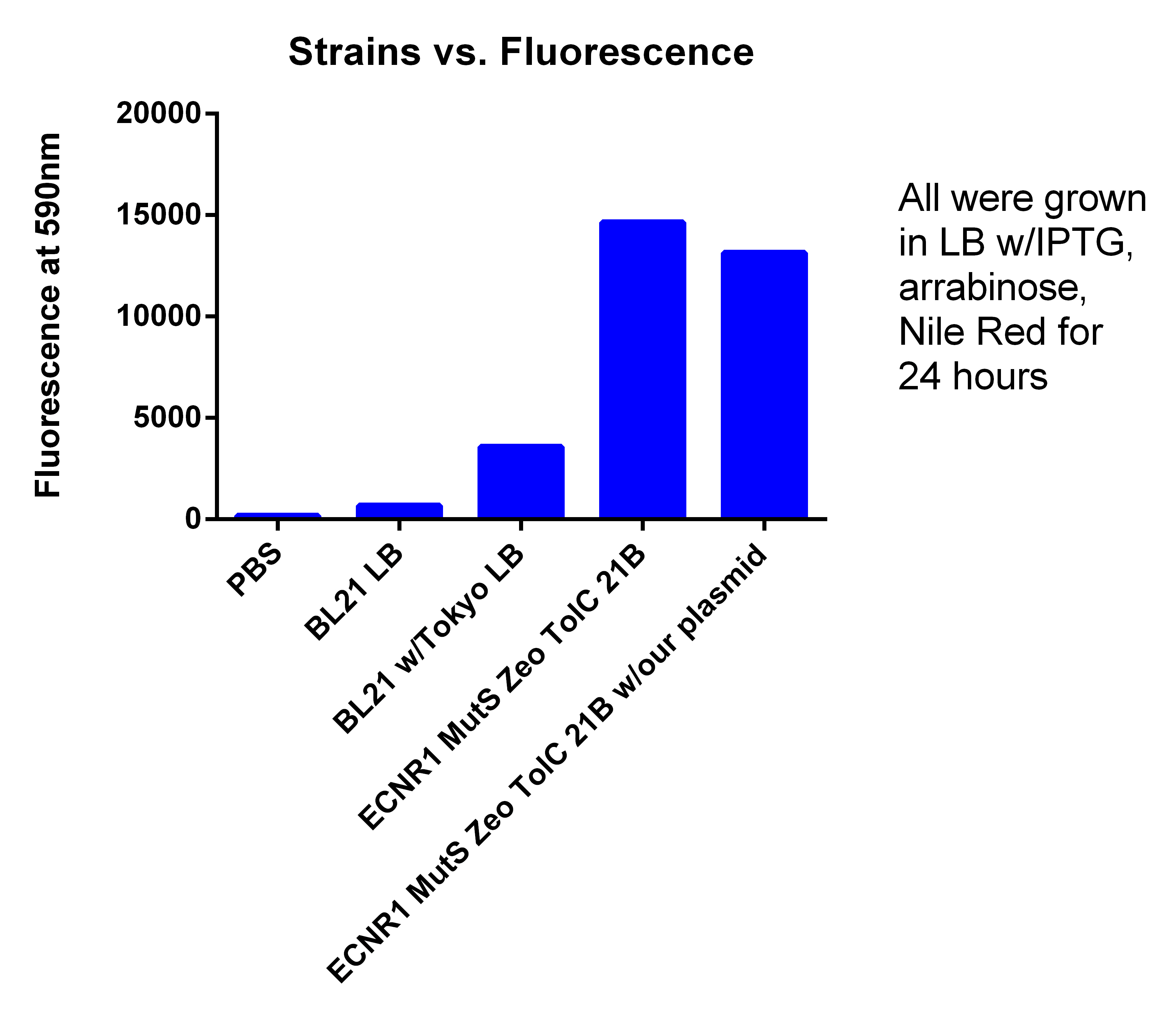
- Tested Andrews positive control
- Procedure, take 1ml of confluent cells, and spun down, washed two times with PBS, and resuspended in 600ul PBS, placed 200ul of PBS + Cells in each well
- PBS = 175.3333333
- BL21 = 679.3333333
- BL21 w/Toyko = 3576.666667
- EcNR1 MutS Zeo TolC 21B = 16632.33333
- EcNR1 MutS Zeo TolC 21B w/our plasmid= 13148
- If you delete the higher number in the EcNR1 MutS Zeo TolC 21B due to the higher OD for that well, the new average is 14649.5
- leans to the fact that we are not producing PLA
- Probably due to LB, not TB with glucose like Korean and Tokyo group did (hopefully my cultures I started today show better results)
- We are not sure why normal EcNR1 is so fluorescent, when it should be just as low as BL21
- Testing the 7 colonies (plus control) from post the Kan selection from Tim
- Tim screened 21B and it appeared to have a band the size of TolC, so not sure how these colonies survived
- Growing each of the 8 strains in 1X SDS to see if they have a functioning tolC
- Also, screening 21B for out construct
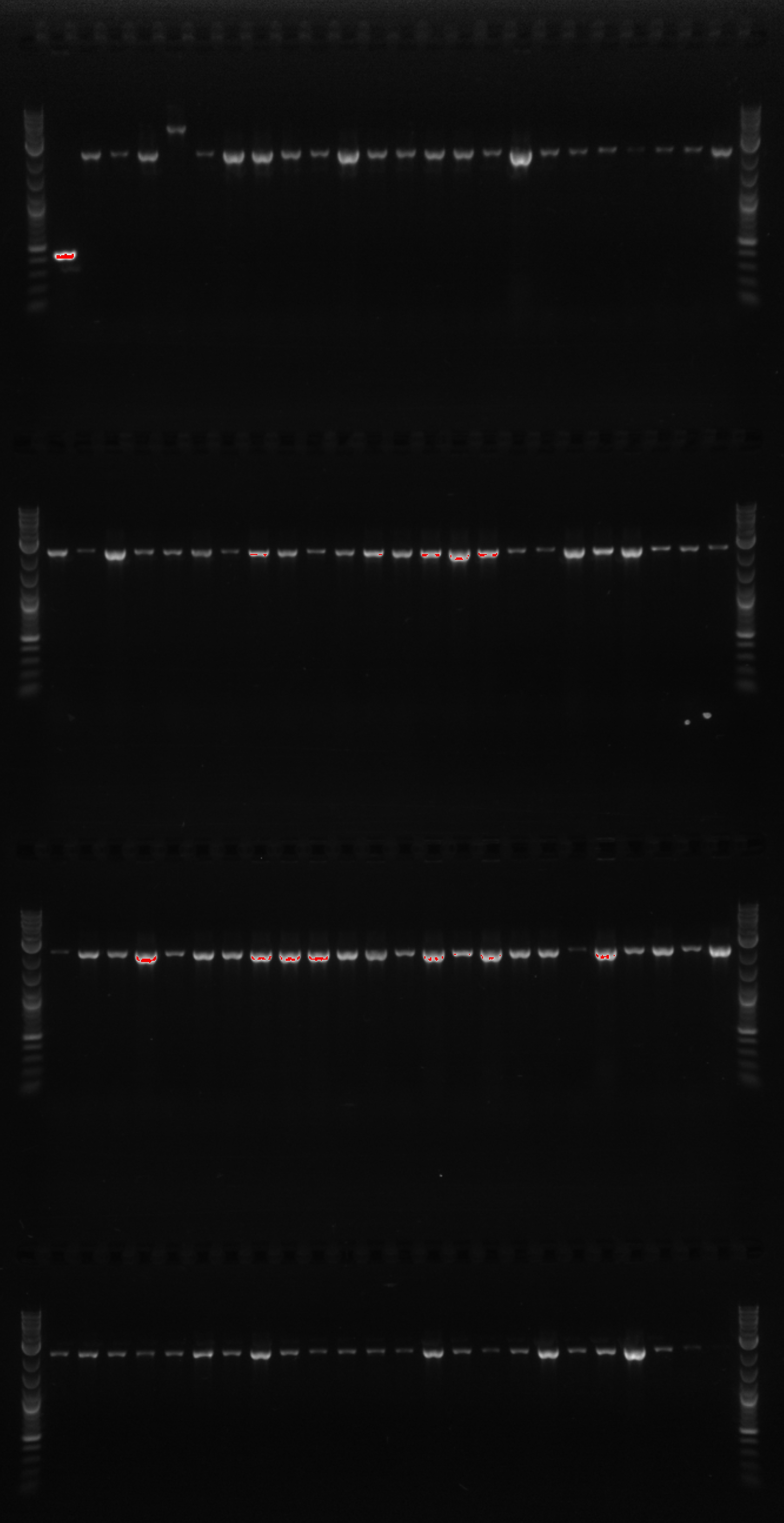
- Using primer 114, and 64 which will create a 1226bp fragment if PHA/PCT are at 21B
- 114 is outside 21B and 64 is internal to our construct
- Using primers 21 and 50 which are both internal to our construct, and should create a 1kb fragment if PHA/PCT is present in the cells
- Using primer 114, and 64 which will create a 1226bp fragment if PHA/PCT are at 21B
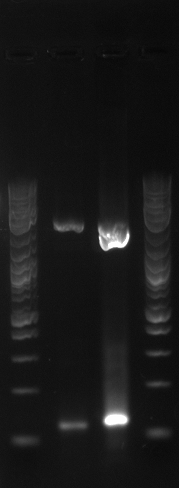
- The lanes on the gel are ladder, PCR1 1, PCR2 1, PCR1 2, PCR2 2, PCR1 3, PCR2 3....ladder)
- Not sure what happened, there appears to be 3-4 bands in the odd lanes which are PCR1 and should have only formed a band if out construct was at 21B
July 25, 2013
- I looked at the sequencing results for the gene Knockouts
- The results are bad, and several are too bad to tell if the gene has been knocked out
- A2 appears to have adhE and pflA knocked out, ackA appears to be functioning, and pflB is too bad to tell
- A3 appears to have none knocked out, adhE appears to be functioning, and ackA, pflA and pflB is too bad to tell
- B4 appears to have adhE (pflA looks bad but still pretty clearly shows knockout), but ackA appears to be functioning, and pflA and pflB is too bad to tell
- The results are bad, and several are too bad to tell if the gene has been knocked out
- Making TB with IPTG, arabinose and Nile red to test fluorescence of both the Tokyo team and ours to see if the cells make more PLA in TB broth
- TB with 250ul of 1mg/ml Nile red, 5ml of 20% arrabinose, and 50ul of 0.1M IPTG
- Made Sprout Presentation for the last class
July 22, 2013-July 24, 2013
- I was at home for my grandfather's funeral
July 19, 2013
- Running negative selection again
- This time on the 3 timed recombineered culture and on the cells grown in SDS
- Backed diluted 1:100 when arrived at 9:30am
- Placed in plate reader at 10:40am
- Does not appear to work, but plated 3 samples at 10^-3 and 10^4 dilutions
- Data here
- Both of the transformations worked
- Started cultures of both and made frozen stocks
- Hopefully the BL21 + PhaC1-A-B1 should be able to produce P(3HB), and like the Toyoko group ground TB is better
- I am making Nile red (0.5ug/ml), glucose (2%), Tb and Lb plates
- Made Lbmin, and followed recipe on TB broth container
- I will add 250ul of the 1ug/ul Nile red, and 50ml of 20% sterile filtered Glucose to make it 2%
- I am making Nile red (0.5ug/ml), glucose (2%), Tb and Lb plates
- Plated normal BL21, BL21+ PhaC1-A-B1 grown in Lb and PhaC1-A-B1 grown in Lb TB in Nile red, glucose and either LB or TB
- Expected results are that normal BL21 should not produce any P(3HB), while the ones with the plasmid should
- Probably the one on the TB plate should make more P(3HB) and thus have a stronger fluorescence when exposed to UV light of the gel reader
- Expected results are that normal BL21 should not produce any P(3HB), while the ones with the plasmid should
July 18, 2013
- Not going to check the negative selection from yesterday because the data suggests that they are both the positive and the negative are growing at the same rate and thus they are really the same population
- So our efficiency must have been so low that they are really almost identical populations
- Our plan is to run cycles with both the one gene (PHA at 21B only) and 2 gene (PHA and PCT at 21)
- Ran two cycles of DsDNA recombination and left the cells growing overnight
- So our efficiency must have been so low that they are really almost identical populations
- Talked to Ryan and he suggested growing up the cells in SDS to kill off any TolC- ones, because the negative (recombineered with water ones) should not grow before 12 hours in the colicin and we are seeing growth
- Grew up ECRN1 tolC 21B mutS Zeo culture in SDS, and running a recombination both with one gene and with two
- Will run negative selection on these two and the 3 round recombinants tomorrow
- Grew up ECRN1 tolC 21B mutS Zeo culture in SDS, and running a recombination both with one gene and with two
- Redoing the transformation from yesterday because Andrew picked the wrong DNA and thus nothing grew on the Chlohpenical plates
- Transformed PZE-GFP into our strain so we will have it and show that the kanR works
- PCR amplify our construct with primers to give homology to KanR
- To amplify our construct PCR1
- F primer 49, R primer PLA_KanR_R (123), length 4.5kb (2.5 minute extension)
- Template is our construct at 1ng/ul
- To amplify KanR PCR2
- F primer Kan_PLA_F (124), KanR_21B_R(125), length 884bp (1 minute extension)
- Template is PZE21 at 1ng/ul
- To amplify our construct PCR1
- PCR our construct to give homology pZE21
- To amplify our construct PCR3
- F primer PLA_PZE_F(126), R primer PLA_PZE_R (127), length 4.5kb (2.5 minute extension)
- Template is our construct at 1ng/ul
- To amplify PZE backbone PCR4
- F primer PZE_PLA_F (128), PZE_PLA_R (129), length 1800bp (1 minute extension)
- Template is PZE21 at 1ng/ul
- To amplify our construct PCR3
- Poured a gel to run these, but didn't have time.
July 17, 2013
- Redoing the Negative selection with both the 2 gene insert (PHA and PCT) and the 1 gene insert (PHA)
- Amounts here
- Placed in shaker at 11 oclock, then placed in plate reader shortly after, and taken out of the plate reader at 1:52
- Plated 10^-3 and 10^-4 dilutions of all 4 positive samples
- All data here
- Picked colonies from the A1 and A5 plates from yesterday
- Running a PCR to screen 21B
- A1: nothing at 21B (430bp)
- A2: TolC at 21B (2000bp)
- A3: OTS from Lexi (3716bp)
- A4-D12:A1 (ancestor), and G1-H12 is also A1 (ancestor) (hopefully makes 4619kb)
- E1: TolC at 21B (2000bp)
- E2-F12:A5 (D8) (hopefully makes 4619bp)
- Running PCR for 2 minute extension time with primers 114 and 115 (430bp fragment if nothing at 21B)
- Amounts here
- Looking for a 4619bp fragment not a 2kb fragment
- Sadly none worked
- A1 is nothing at 21B (400bp), A3 is TolC, and A5 is Lexi at 3.7kb
- Backup Plans
- Put our construct in a PZE plasmid so can do positive selection with Kan resistance
- Amplify our construct and add Kan resistance via gibson assembly
- Take KanR from PZE plasmid
- Primers have been ordered for these two options
Options to get genes in E. Coli:
- Screen the negative selection from today
- Would need to check the size of the two bands but probably use a primer that is in the middle of the fragment (either tolC or our construct or both)
- Run more cycles of both genes and only one gene
- Would still have to use negative selection
- Put our genes on a PZE21 and use positive selection with Kanamycin
- Add kanR to the end of our fragment and integrate into the genome
- Add resistance on to PHA only, and the need to add another resistance on other gene
July 16, 2013
- Back diluted cells for negative control
- Here is new protocol
- Shook in the high speed shaker next to water bath in 204 for 4.5 hours, and the put in plate reader to monitor growth curve
- Here is the first OD measurement at 4.5 hours
- Growing both ECNR1 without TolC, and our strain (ECNR1 tolC 21B, MutSZeo) in SDS (1.5ul of 2000X)
- Only our strain should grow
- Control to make sure that our strain really does have TolC because negative selection isn't working well
- Also, we could use this for the next negative selection to make sure any contamination or other colonies are dead
- here is picture from next morning and only our strain grew, which means TolC is active
- Also, pursuing the back up plan, of inserting just PHA to 21B, then tolC at frd, and then PCT at frd to replace tolC
- Grew up the EcRN1 mutS zeo for the first DsDNA recombination
- Using the PHA gene with homology to 21B at both ends, a 105.45ng/ul
- Will use 1ul of the PHA in 49ul of water as DsDNA and 50ul water for negative control
- Grew up the EcRN1 mutS zeo for the first DsDNA recombination
- Preform the recombination and cells are growing upstairs in the incubator ovrenight
- Also, tried a long shot
- Plated 10^-3 of the positive recombineered ancestor strain on IPTG, arrabinose, Nile Red plates
- Will test on blue light in the slight chance that one is producing PLA and fluorescent, and will pick and try to isolate that colony
- Sequencing results from keck came here
- I aligned all of the sequences with the ape file in Geneious
- Here is the file
July 15, 2013
- Although we picked colonies, after our meeting with Farren we don't think the negative selection worked
- I would redo it, but don't have enough time today, so will start it again first thing tomorrow
- I back diluted the recombineered cultures from Friday and will use those tomorrow
- We are simplifying and getting rid of the veconmycin, and only growing 1:100
- We are growing the cells in the high speed shaker in 204 rather than the shaking incubator
- Here is new protocol
- Pursued the plan to put the genes in separate
- PHA at 21B, and PCT at FRD
- Diluted each gene to 1ng/ul
- PHA: primer 49, 101, 2383bp
- PCT: primer 102, 103, 2006bp
- Diluted each gene to 1ng/ul
- PHA at 21B, and PCT at FRD
- Ran at 60 degrees annealing and 1:15 extension time
- Ladder, 4ul PHA 21B, rest PHA 21B, 4ul PCT frd, rest PCT frd, ladder
- Gel purified and will nanodrop tomorrow
- PHA at 105.45ng/ul
- PCT at 91.72ng/ul
- Attempting to figure out if we can detect Nile red using any fluorescence reader we already have
- Nile Red in DMSO was fluorescent under the Blue light in 204, and less fluorescent under Ethanol, and not visibly fluorescent under water
July 14, 2013
- Redo negative Selection with re-recombineered strains from Friday
- Record when the plate goes in the incubator/shaker.
- Check growth every 20-30 mins starting at 5 hours (The negative selection will take 6-8 hours)
- When there is growth in your wells recombined with your DNA, but not with the water, stop the negative selection by removing the plate.
- Bring it to the bench, do dilutions, and plate on LB - dilutions will vary depending on how dense the well looks. If you are unsure, do more dilutions than less. You'll need single colonies
- Placed in incubator at 12:30pm
- Blue is wrong, and orange were plated in 10^-3 and 10^-4 dilutions
- Going to sequence our construct, using sequencing primers and primers we used to amplify since the sequencing primers were designed for 21B so can't use the first and last one
- Fragment 1: 49 and 12, 949bp, n/a F, Z R
- Fragment 2: 13 and 14, 1105bp, A F, Z R
- Fragment 3: 15 and 16, 1100bp, B F, Z R
- Fragment 4: 17 and 18, 1067bp, A F, Z R
- Fragment 5: 19 and 61, 658bp, A F, n/a R
- 12-19 are sequencing primers, while 49 and 61 are just normal primers
- 58 degrees for 30 second extension time for all 5
- Going to run a small amount on a gel to make sure there is band at the right length, and then PCR purify the DNA, nanodrop and prepare for sequencing
- Gel is Ladder, frag 1, frag 2, frag 3, frag 4, frag 5, ladder
- Submitted 8 samples for sequencing
July 12, 2013
- Picked colonies, 48 from each of the three strains that were plated (ancestor, A9 and A11)
- Picked into 20ul water, and placed 1.5ul of this water into a new plate with 150ul LB and put in incubator
- Now will run PCR with 1ul from the water plate as template
- Running a PCR to see if our construct (4.5kb) or TolC (1.5kb) is at 21B, will use sequencing primer 1 and 10
- Forward GTAAAACGACGGCCAGcaccagcgataacgcttcacg (64.88)
- Reverse CAGGAAACAGCTATGACccggatgcctcgaaagataagc (64.07)
- Amounts Here
- Setup of gel:
- First row is Ancestor before MAGE: ladder, A1, B1......C1, D1.....ladder
- Second row is A9: ladder, A1, B1......ladder, C1, D1.....ladder
- Third row is A11: ladder, A1, B1......ladder, C1, D1.....ladder
- The non-transformed ancestor is in the second well of row 1 and 3
- None of them worked (sadness)
- Redid Double stranded recombineering with twice as much of our construct (4ul in 200ul)
- Performed the recombineering on the Ancestor, A9, A11 and D8
- Have a positive and negative control and resuspended in 1ml and left growing in the incubator
July 11, 2013
- Negative selection, use a flat bottom 96 well plate
- Add LB first, then vancomycin (to half), then colicin quickly and keep on ice (in -80 freezer)
- Then add cells in 1:100
- Grow for 5 hours, then eyeball---need one that has nothing in negative control, and cells in the positive recombineer
- Let cells grow for 5 hours, and 5/8 showed expected results
- Plated A1 (ancestor) in 10^-3 and 10^-4 dilutions, Plated A2 (ancestor) in 10^-3 and 10^-4 dilutions, Plated A3 (culture A9) in 10^-4 and 10^-5 dilutions, Plated A5 (A11) in 10^-3 and 10^-4 dilutions, Plated A6 (A11) in 10^-3 and 10^-4 dilutions,
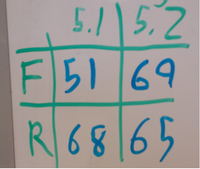
- Designed and ordered lots of primers
- Made all of the universal primers the same as on the Keck website, so they should all work
- Re-ordered primers to sequence both genes at 21B with correct universal, so all ones we currently have are trash
- All forward sequencing primers should have: TGTAAAACGACGGCCAGT
- All reverse sequencing primers should have: TCACACAGGAAACAGCTATGAC
- Re-ordered primers to sequence both genes at 21B with correct universal, so all ones we currently have are trash
July 10, 2013
- Re-organized the DsDNA so important fragments are in a new box called DsDNA2
- Run PCR from yesterday on a Gel
- Ladder, 4ul PCR 1 square, rest PCR 1 square, 4ul PCR 2 square, rest PCR 2 square, 4ul PCR 1 round, rest PCR 1 round, 4ul PCR 2 round, rest PCR 2 round, ladder, 4ul TolC, rest TolC, ladder
- PCR should all be 4540bp long, and tolC should be about 1.5kb
- ALL PCRs of the full gene appeared to have worked
- The TolC has a band at the right length (band at the bottom is most likely primer dimer)
- PCR should all be 4540bp long, and tolC should be about 1.5kb
- Gel purified and Nanodroped PHA-PCT_21B is 246.889ng/ul, TolC with homology to frd 20.716ng/ul
- Recombineer our construct into our strain
- Will do 4 recombineering (plus a negative control for each)
- ECNR1 MutS Zeo tolC 21B, ECNR1 MutS Zeo tolC 21B 5x MAGE A9, ECNR1 MutS Zeo tolC 21B 5x MAGE A11, ECNR1 MutS Zeo tolC 21B 5x MAGE D8
- Will add 2ul of our DNA at (246.889ng/ul) to 200ul water to get a concentration of 123ng in 50ul of H2O
- Will do 4 recombineering (plus a negative control for each)
- Instructions from Natalie
- Start cultures of cells to recombineer
- Set up recombineering stuff. This will be the same as MAGE, i.e. recovery culture, tubes on ice, etc., EXCEPT: instead of 1uM for DNA, use 100+ng of DNA in 50uL of water. ALWAYS do negative control with water, you'll need it for negative selection, one for every strain you use (so that'll effectively double the number of samples).
- When cells are mid-log, induce (just like MAGE)
- Follow MAGE protocol
- After electroporation, let cells recover in culture tube in shaking incubator overnight.
- Left the 8 cultures tubes (a positive and negative control for each of the 4 strains) in incubator overnight
July 9, 2013
- Ran PCRs from yesterday on a gel
- Ladder,4ul PCR 1, rest PCR 1, 4ul PCR 2, rest PCR 2, 4ul PCR 31, rest PCR 3, ladder
- PCR 1 and 2 are both 1966bp, while PCR 3 is 995bp
- Gel purified and nanodropped
- PCR 1 3.78ng/ul
- PCR 2 36.8ng/ul
- PCR 1 22.7ng/ul
- Ran the gel purified PCR product on a new gel in small quantities to see exactly where they run
- Ladder, 2ul PCR2, 6ul PCR 1, 4ul PCR3, ladder
- Gisbon assembly both genes
- Run PCR of the Gibson assemblies
- Primer 49 and 61 (lower primer has Tm of 80 degrees) fragment 4289bp long
- 2:30 minute extension and 65 degrees
- Primer 49 and 61 (lower primer has Tm of 80 degrees) fragment 4289bp long
- Amplify TolC with homology to frd complex
- TolC from Natalie plasmid
- Primers 79 and 80, 1.5kb fragment (45 seconds) at 58 degrees
- TolC from Natalie plasmid
- Designed and ordered primers to put both Biobricks (for export system) into PZE21 plasmid
- Ordered MAGE oligos
July 8, 2013
- Made new Xylene Cyanol loading dye
- Mixed Ladder with loading dye
- Ordered two primers to amplify tolC with homology to frd genes
- Ordered new reverse primer to amplify the gibson assembly of the first gene in order give it homology to 21B at both ends in order to put each gene in separate
- Pictures of the zeo control plates
- Took a big and small colony off the transformation and plated it both on Zeo and normal plates
- Since both formed big and small colonies, we concluded that it is just an effect of the Zeo, and there are not two seperate genoytpes at place
- PCR amplified fragment 7 with new primers (72 and 73) to increase the overhang and thus hopefully fix the problem with the Gibson assembly
- 58 degrees and 30 second extension time
- Ran PCR product on a gel (looking for a 560bp fragment)
- Ran a gibson assembly of the second gene
- PCR amplified all Gibson assemblies
- Gibson #1:Primer 51 and 61, 67 degrees, 1 minute extension (fragment 1966bp)
- Gibson #2:Primer 51 and 61, 67 degrees, 1 minute extension (fragment 1966bp)
- Gibson #3:Primer 72 and 61, 68 degrees, 30 second extension (fragment 995bp)
July 7, 2013
- Took another look at the second gene fragments, starting over
- Here is the ape file
- No glaring mistakes that would cause the gibson to fail
- However, the overhang temperature between fragment 7 and 8 is 51 degrees which is boarderline too low, this probably isn't helping, plus this has a hairpin
- The new primers we already order should fix this problem
- Also, the temperature between the first and second gene is only 53 degrees, so I designed a new reverse primer for terminator of PHA, and a new forward for 5.1 to increase the overlap if they won't properly assembly
- 5.1F cgcgttatgcaggTCCCTATCAGTGATAGAGATTGACATCC
- T1R CTCTATCACTGATAGGGAcctgcataacgcgaagtaatcttttc
July 5, 2013
- Made and ordered sequencing primers
- Streaked the recombineered Ecnr1 with MutS Zeo onto 2 new plates as a control
- Streaked a big colony onto a normal and zeo plate
- Streaked a small colony onto a normal and zeo plate
- Ran the PCR reactions from yesterday on a gel
- Lanes are Ladder, 4ul 0% DMSO Gibson, rest 0% DMSO Gibson, 4ul 2% DMSO Gibson, rest 2% DMSO Gibson, 4ul 5% DMSO Gibson, rest 5% DMSO Gibson, ladder, 4ul Part 1 Gibson, rest Part 1 Gibson, 4ul Part 2 Gibson, rest Part 2 Gibson, ladder
- Gibsoning the whole gene doesn't appear to be working but part 1 (which has 5.1, 5.2 and 6) does appear to be accurate
- This means there is probably a problem with connecting 6 to 7, 7 to 8, 8 to T2.
- I ran 4 more gibson assemblies to try and determine which overhang is the problem
- Here are the amounts
- PCR amplified all 4 gibson assemblies at 63 degrees annealing and 30 second extension time
- 6 and 7 using primers 27 and 30 (fragment 1020bp)
- 7 and 8 using primers 29 and 52 (fragment 699bp)
- 8 and T2 using primers 31 and 61 (fragment 466bp)
- 7, 8, T2 using primers 29 and 61 (fragment 956bp)
- Ran PCR on a Gel
- Lanes are Ladder, 4ul 6 and 7, rest 6 and 7, 4ul 7 and 8, rest 7 and 8, 4ul 8 and T2, rest 8 and T2, 4ul 7, 8 and T2, rest 7, 8, and T2, ladder
- It appears that only 8 to T1 is working
- Running the assemblies on a new gel to see if the PCR is the problem or the assembly
- ladder, 6-7, 7-8, 8T1, 7-8-T1
- Gel does not have that much helpful information
- The assembly of 6 and 7 appears to have made a band around 700 which is very strange
- Gel purified this band and will try and PCR both fragments (6 and 7) off of it
- The assembly of 6 and 7 appears to have made a band around 700 which is very strange
- Redesigned the primers on fragment 7 with increase the overhangs in attempt to fix this gibson assembly
- Both ends of this fragments were hairpins which could be the problem
- New 7 reverse: ataagtgcaaaccatcttccttcaaaaggaatacgcatctttctgtaatataagttac
- New 7 forward primer: cattgaattagaaaaagatgttgctgtaaatttaggtgttggtgcgcctgaatatgtagc
- Both ends of this fragments were hairpins which could be the problem
July 4, 2013
- Run the PCR from yesterday (Gibson assembly of the PCT gene parts, with homology to PHA and 21B) on a gel
- Looking for a fragment of size 1956
- Ladder, 4ul PCR1, rest PCR1, 4ul PCR2, rest PCR2, nothing, ladder
- Got double bands so did not work properly
- Most likely it either didn't insert on of the 6 fragments or inserted one twice
- Ran 5 more gibson assemblies in order to hopefully get one that works
- Tried different amounts of DMSO, and breaking the gibson into two smaller (3 fragment) assemblies
- 0% DMSO, 2% DMSO, 5%DMSO
- Amounts here
- Tried different amounts of DMSO, and breaking the gibson into two smaller (3 fragment) assemblies
- Ran PCR of the 5 Gibson assemblies
- All 3 assemblies with all fragments were run using primers 51 and 61 at 68 degrees and looking for a 1966 fragment
- The first part was run with primers 51 and 28 run at 67 degrees for 1 minute extension and is 1020bp long
- The second part was run with primers 29 and 61 run at 67 degrees for 30 second extension and is 966bp long
July 3, 2013
- Going to PCR fragment 5 in two parts in order to fix the deletion mutation (with new primers)
- Run the PCR at 58 degrees annealing temp
- Run for 15 seconds
- Ran two samples for 5.1 and 5.2 with two different dilutions of the minigene (one in green container, one in purple, just in case)
- Run on gel and look for bands
- PLA 5.1 is 208bp and PLA 5.2 is 291
- Gel purfiy and nanodrop (concentrations 37.5 and 28.5)
- Wells are Ladder, 4ul PLA 5.1 green, rest PLA 5.1 green, 4ul PLA 5.1 purple, rest PLA 5.1 purple, ladder, 4ul PLA 5.2 green, rest PLA 5.2 green, 4ul PLA 5.2 purple, rest PLA 5.2 purple, ladder
- Used two different dilutions (both 1 ng/ul but made different days), just in case
- Gibson assembly PCT gene (PLA5-8 + term2)
- PCR amplify the fragment with primers 51 and 61
- Will have homology to the end of the PHA fragment, and 21B
- There temps are both 70 so could run pretty high
- Looking for a fragment of size 1956
July 2, 2013
- Redid PCR of the gibson assembly because may have used the wrong reverse primer and also need to run it for 1:15 because it is 2.5kb
- Use primers 49 and 59
- Here is the latest list for the primers to combine both genes together and insert in 21B
- Does not include fixing the missing base in fragment 5
- Run the PCR of the first gene on gel
- The lanes are ladder, PCR from 7-1, PCR from 7-1, Ladder, PCR from 7-2, PCR from 7-2, Ladder
- As expected I used the wrong reverse primer (should have used 59 but used 58), so the PCR from 7-1 did not work
- The PCR from 7-2 appears to work but I forgot to run 4uL to get a more accurate reading
- Looking for a band 2584bp
- Gel purified and nano droped the fragment (concentration 162.462ng/ul)
- Ran a gel with ladder, PLA1, PLA2, PLA3, PLA4, Terminator 1, Gibson all all 5 previous fragments, ladder
- It appears that the gibson assembly worked because PLA1-PLA4 all ran around 500bp, terminator 1 is 284bp, and the assembly is 2584bp
- Ordered two primers so if we want to use gBlock 5 and PCR the promoter off of the minigene #5 using these two new primers plus the original two primers for PLA 5
- Transformed minigene (PLA #5) into XL1 Blue and grew on Kan plates
July 1, 2013
- Grew both the positive recombineered strain and before the recombineer in 3ml of Zeo as a control
- This is a control because the colonies on the Zeo plates were different sizes which shouldn't have happened
- Control worked, only the recombineered strain was able to grow in Zeo medium
- The top tube contains the ancestral ECNR1 and no visible growth after 17 hours
- The bottom tube contains the recombineered ECNR1 Muts:Zeo and has very visible growth after 17 hours
- Ran a PCR to amplify fragments 2 and 8
- Ran a gibson assembly of PLA fragments 1-4 with the first terminator for one hour at 50C
- Has homology to 21B and then to PCT
- Amounts added here
- PCR the result of the Gibson with primer 49 and 58 (it should have been 59 so will redo just in case)
June 28, 2013
- Only the recombineer plates grew colonies
- Picked 47 colonies and grew on 98 well plate (A1 was ancestor strain ECNR)
- PCR tested for MutS with primers 157 and 158 from Ryan
- See if band is either MutS (2.6kb) or Zeo (0.8kb)
- All except 1 well appeared to be the correct length (the primers add about 0.5kb)
- Streaked a Zeo plate using well A2
June 27, 2013
- Performed recombination knocking out MutS with zeo
- Strain was ECNR1 with tolC and pbad at 21B from Jaymin
- Zeo doubled stranded DNA from Ryan
- Plated both recombineer and negative control on zeo plates
June 26, 2013
- Attempted a negative control assay but unable to detect PLA uptake using the blue light filter in room 204
- Thought it may work because Nile red is fluorescent at roughly the same wavelength as Gel green
- As soon as PLA in Acetone was placed in LB or on a plate it came out of solution, so can't test assay
- Probably should just order the Pseudomonas
June 24, 2013
- Ran another PCR of fragment #3
- Made new working stocks of both the primers and the template DNA
- Raised the annealing temperature
- Well 1 and 9 Ladder, then temperatures increase from 58, 58.7, 60, 62, 64.4, 66.4, 67.5, 68
- Since all appeared to work, we took 3 (58 degrees, 62, and 67.5 and ran the rest of the PCR mix)
- Gel purified
June 20, 2013
- PCR amplified and gel purified the 6/8 g-blocks from IDT
- PLA fragments 1,3,4,6,7,8 were ran in PCR using primers previously ordered
| Well Number | Sample |
|---|---|
| 1 | Ladder |
| 2 | 4ul PLA 1 (500bp) |
| 3 | Rest PLA 1 (500bp) |
| 4 | 4ul PLA 3 (500bp) |
| 5 | Rest PLA 3 (500bp) |
| 6 | 4ul PLA 4 (496bp) |
| 7 | Rest PLA 4 (496bp) |
| 8 | 4ul PLA 6 (500bp) |
| 9 | Rest PLA 6 (500bp) |
| 10 | 4ul PLA 7 (500bp) |
| 11 | Rest PLA 7 (500bp) |
| 12 | 4ul PLA 8 (173bp) |
| 13 | Rest PLA 8 (173bp) |
| 14 | Ladder |
| PLA part | Concentration (ng/µL) |
|---|---|
| PLA 1 | 38.177 |
| PLA 3 | 6.85 |
| PLA 4 | 28.793 |
| PLA 6 | 23.734 |
| PLA 7 | 53.491 |
| PLA 8 | 17.059 |
- Something seems to have gone wrong with the 3rd PLA fragment
June 14, 2013
- Took pictures of the second cycle of MAGE practice
June 12, 2013
- Made graphs of negative controls
- Replated MAGE colonies and they worked (initial plates went bad, due to light exposure)
June 11, 2013
- Ran another attempt at negative control
| Sample | Well #1 Fluorescence at ex. 530, em. 590 | Well #2 Fluorescence at ex. 530, em. 590 | Well #2 Fluorescence at ex. 530, em. 590 | Average Fluorescence |
|---|---|---|---|---|
| 200ul Water | 182 | 176 | 179 | 179 |
| 200ul Water with 10uL Nile Red* | 220 | 217 | 225 | 220.67 |
| 200ul PBS | 182 | 183 | 188 | 184.33 |
| 200ul PBS 10uL Nile Red | 222 | 240 | 216 | 226 |
| 200ul 60% lactate | 294 | 325 | 327 | 315.33 |
| 200ul 60% lactate with 10uL Nile Red | 334 | 321 | 326 | 327 |
| 200ul LB | 567 | 565 | 561 | 564.33 |
| 200ul LB with 10uL Nile Red | 611 | 624 | 649 | 628 |
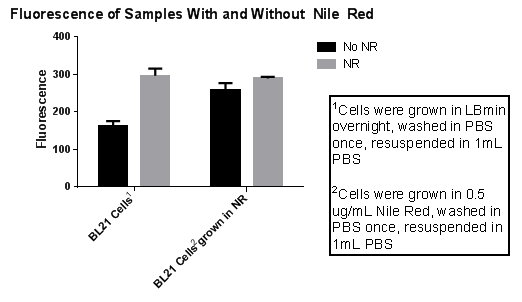
| BL21 Cells** | 160 | 177 | 150 | 162.33 |
| BL21 Cells with 10uL of Nile Red added** | 296 | 278 | 316 | 296.67 |
| BL21 Cells grown in Nile red*** | 239 | 273 | 266 | 259.33 |
| BL21 Cells grown in Nile red with 10uL of Nile Red added*** | 293 | 291 | 285 | 289.67 |
| 200ul Acetone | 67 | 70 | 84 | 73.67 |
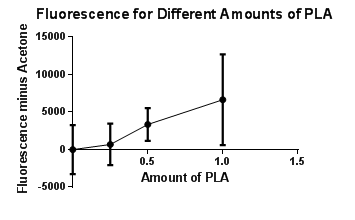
| 200ul Acetone with 10uL Nile Red | 22162 | 26722 | 28509 | 25797.67 |
| 100uL PLA, 100uL Acetone with 10uL Nile Red | 34223 | 37419 | 25718 | 32453.33 |
| 50uL PLA, 150uL Acetone with 10uL Nile Red | 26666 | 30100 | 30719 | 29161.67 |
| 25uL PLA, 175uL Acetone with 10uL Nile Red | 23317 | 27718 | 28418 | 26484.33 |
| *Nile red at a concentration of 10ug/ml | ||||
| **Cells were grown in LBmin overnight, spun, supernatant removed, added 1X PBS, spun, supernatant removed, resuspended in 1mL PBS | ||||
| ***Cells were grown in 0.5 ug/mL Nile Red, spun, supernatant removed, added 1X PBS, spun, supernatant removed, resuspended in 1mL PBS |
June 7, 2013
- Ran another attempt at negative control
| Sample | Well #1 Fluorescence at ex. 530, em. 590 | Well #2 Fluorescence at ex. 530, em. 590 | Average Fluorescence |
|---|---|---|---|
| 200ul Water no Nile Red | 156 | 156 | |
| 200ul PBS no Nile Red | 169 | 169 | |
| 200ul Water with 10uL Nile Red* | 116 | 194 | 155 |
| 200ul Acetone with 10uL Nile Red | 12843 | 11559 (Probably less than 200uL Acetone) | 12201 |
| 200ul 60% lactate with 10uL Nile Red | 243 | 297 | 270 |
| 200ul LB with 10uL Nile Red | 434 | 460 | 447 |
| 200ul PBS (1X) with 10uL Nile Red | 176 | 128 | 152 |
| 200ul PLA with 10uL Nile Red | 10079 | 8695 | 9387 |
| 100uL PLA, 100uL Acetone with 10uL Nile Red | 11557 | 9589 | 10573 |
| 50uL PLA, 150uL Acetone with 10uL Nile Red | 13210 | 12931 | 13070.5 |
| 25uL PLA, 175uL Acetone with 10uL Nile Red | 14994 | 18019 | 16506.5 |
| Chunk of PLA soaked in Nile red for 6 hours | 413 | 413 | |
| Supernatant of chunk of PLA soaked in Nile red for 6 hours | 160 | 190 | 175 |
| BL21 Cells** grown in Nile Red for 6 hours | 255 | 254 | 254.5 |
| BL21 Cells grown in Nile Red for 6 hours with 25uL of Nile Red in well | 389 | 369 | 379 |
| *Nile red at a concentration of 10ug/ml | |||
| **Cells were grown in Nile red for 6 hours, spun, supernatant removed, added 1X PBS, spun, supernatant removed, resuspended in 1mL PBS |
June 6, 2013
- Tried dissolving PLA in 2-Propanol: no PLA appeared to dissolve
- Ran a negative control with the supernatant from test tube A (not all PLA dissolved)
- Data below (all had 15uL of nile red (10ug/ml) except water)
| Sample | Fluorescence at ex. 530, em. 590 |
|---|---|
| Water | 185 |
| Acetone | 17746 |
| Lactate 60% wt | 379 |
| PLA in Acetone | 20652 |
| 0.5 PLA | 20088 |
| 0.25 PLA | 22245 |
| 0.125 PLA | 22480 |
June 5, 2013
- Attempting to dissolve PLA in acetone to collect data for Sackler presentation
- Unsuccessful when attempting to placed PLA in acetone in glass beaker with stir bar
- 5mL tubes were eaten by acetone
- Cut PLA and added roughly half a pellet to 1.5ml eppendorf tubes, left in sonicator
- Attempted to crush PLA with hammer and had difficulties, added small amount to 1 ml Acetone and left in sonicator
| Sample | Amount of PLA | Solvent |
|---|---|---|
| A | 0.0160g | 1ml Acetone |
| B | 0.0131g | 1ml Acetone |
| C | 0.0120g | 1ml Acetone |
| D | 0.0142g | 1ml Acetone |
| E | 0.0139g | 1ml Acetone |
| F | 0.0015g | 1ml Acetone |
| G | 0.0017g | 1ml 2-Propanol |
 "
"