Team:BYU Provo/Notebook/LargePhage/Springexp/Period3/Dailylog
From 2013.igem.org
Brymerr921 (Talk | contribs) (→5/29/13) |
Brymerr921 (Talk | contribs) (→5/27/13) |
||
| Line 4: | Line 4: | ||
===5/27/13=== | ===5/27/13=== | ||
| - | |||
===5/29/13=== | ===5/29/13=== | ||
Revision as of 21:25, 5 June 2013
Contents |
5/27/13
5/29/13
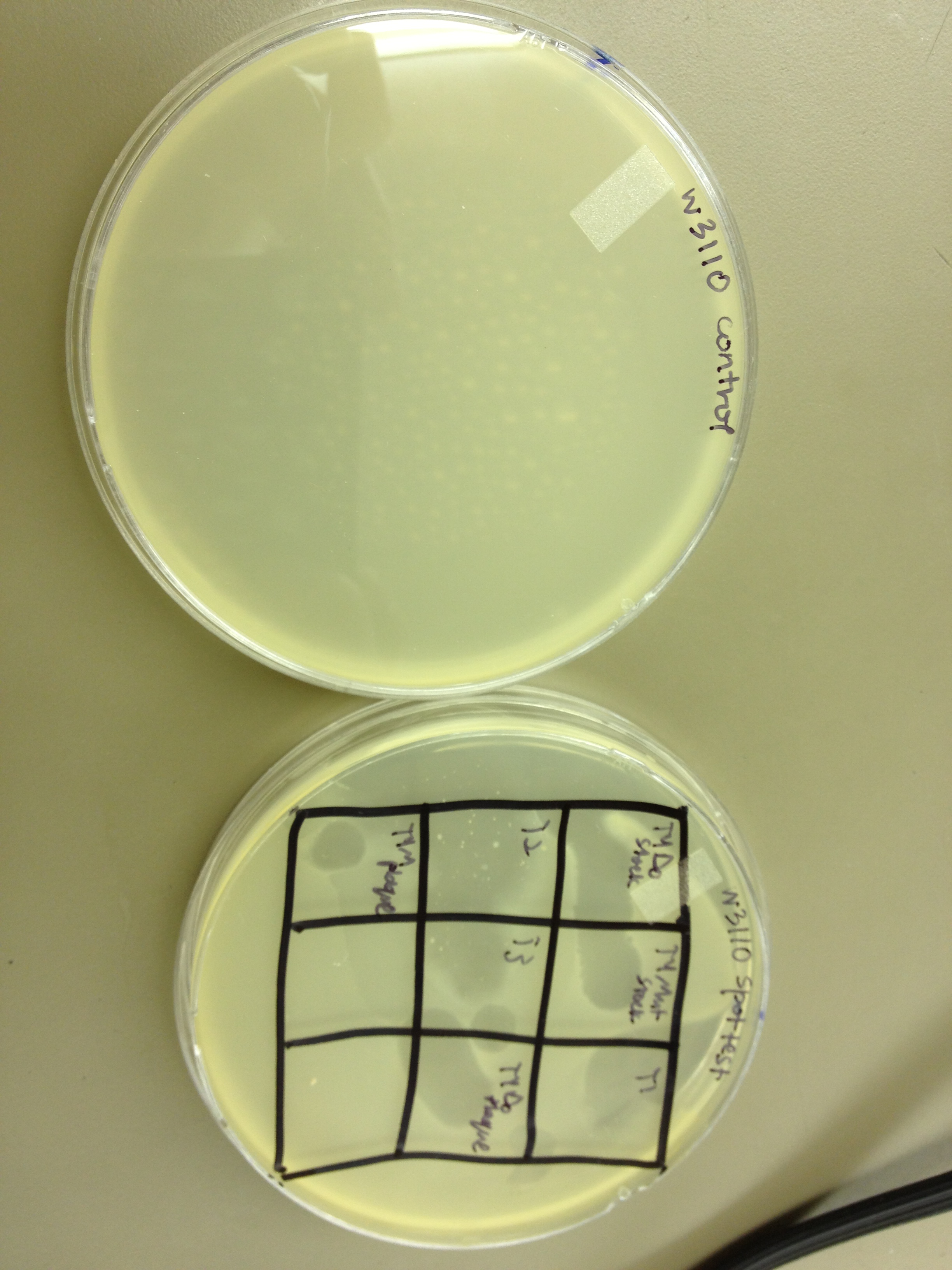
Today we wanted to see if we can transfer our current experiment from using E. coli W3110 as the host to E. coli B as the host. We set up multiple experiments.
1. We spot tested the following phage samples on W3110 and B: T1, T2, T3, T4Do stock, T4 mutated stock, T4-UV one-plaque plate harvest, T4-UV-mutated one-plaque plate harvest.
2. We did a dilution series on the T1 stock to see how concentrated it is. We will need to know the PFUs in order to grow a successful liquid culture and mass produce it for our large phage amplification procedure.
3. We also infected two samples of E. coli B, each with 10 uL of 10^-6 T4Do to observe plaque formation.
We archived two cryo-tubes of E. coli B by mixing 600 uL each of overnight E. coli B and 40% glycerol. These two tubes will be stored at -80C.
5/31/13
I took out the plates from last time's experiments.
1. The spot test showed that all seven samples cleared both E. coli B and W3110.
2. However, our dilution series for T1 did not work. All of the plates appeared to show only bacterial lawns. The plates were not observed after a 24 hour incubation, but 48 hours later there were only lawns on all the plates.
3. The 10^-6 T4Do plates produced excellent plaques, confirming that T4Do infects E. coli B.
Also, today Jade taught me how to set up the website- so I worked on that.
June
6/3/13
KS- I spent most of today getting the website in order and organized for future experiments to be entered in easily!
6/5/13
6/7/13
 "
"