Template:Kyoto/Notebook/Aug 21
From 2013.igem.org
(Difference between revisions)
(→Electrophoresis) |
(→Gel Extraction) |
||
| Line 310: | Line 310: | ||
|4 | |4 | ||
|} | |} | ||
| - | [[File:Igku_Aug21_Gel_Extraction(DT_EcoRI+XbaI_12,5μL①)before.jpg]] | + | [[File:Igku_Aug21_Gel_Extraction(DT_EcoRI+XbaI_12,5μL①)before.jpg]] |
| - | [[File:Igku_Aug21_Gel_Extraction(DT_EcoRI+XbaI_12,5μL②)after.jpg]] | + | |
| + | [[File:Igku_Aug21_Gel_Extraction(DT_EcoRI+XbaI_12,5μL②)after.jpg]] | ||
</div> | </div> | ||
Revision as of 03:52, 26 September 2013
Contents |
Aug 21
Plusgrow Medium
- material
- 8/17 Plusgrow medium 50ml
- 7/22 5000x Ampicillin 10µl
- Measuring materials and putting them in a 50ml tube
Liquid culture
| Sample | medium |
|---|---|
| 8/20 tRNA-spinach -(1) | 8/21 Plusgrow medium(+Amp) |
| 8/20 tRNA-spinach -(2) | 8/21 Plusgrow medium(+Amp) |
| 8/20 TetR-aptamer 12_P(1076) -(1) | 8/21 Plusgrow medium(+Amp) |
| 8/20 TetR-aptamer 12_P(1076) -(2) | 8/21 Plusgrow medium(+Amp) |
| 8/20 TetR-aptamer 12_1R(113) -(1) | 8/21 Plusgrow medium(+Amp) |
| 8/20 TetR-aptamer 12_1R(113) -(2) | 8/21 Plusgrow medium(+Amp) |
| 8/20 TetR-aptamer 12_1M(105) -(1) | 8/21 Plusgrow medium(+Amp) |
| 8/20 TetR-aptamer 12_1M(105) -(2) | 8/21 Plusgrow medium(+Amp) |
| 8/20 PT181 attenuator -(1) | 8/21 Plusgrow medium(+Amp) |
| 8/20 PT181 attenuator -(2) | 8/21 Plusgrow medium(+Amp) |
| 8/20 Fusion1 attenuator -(1) | 8/21 Plusgrow medium(+Amp) |
| 8/20 Fusion1 attenuator -(2) | 8/21 Plusgrow medium(+Amp) |
| 8/20 Fusion3m2 attenuator -(1) | 8/21 Plusgrow medium(+Amp) |
| 8/20 Fusion3m2 attenuator -(2) | 8/21 Plusgrow medium(+Amp) |
| 8/20 PT181 antisense -(1) | 8/21 Plusgrow medium(+Amp) |
| 8/20 PT181 antisense -(2) | 8/21 Plusgrow medium(+Amp) |
| 8/20 Fusion1 antisense -(1) | 8/21 Plusgrow medium(+Amp) |
| 8/20 Fusion1 antisense -(2) | 8/21 Plusgrow medium(+Amp) |
| 8/20 Fusion6 antisense -(1) | 8/21 Plusgrow medium(+Amp) |
| 8/20 Fusion6 antisense -(2) | 8/21 Plusgrow medium(+Amp) |
- incubate 37°C 10hour
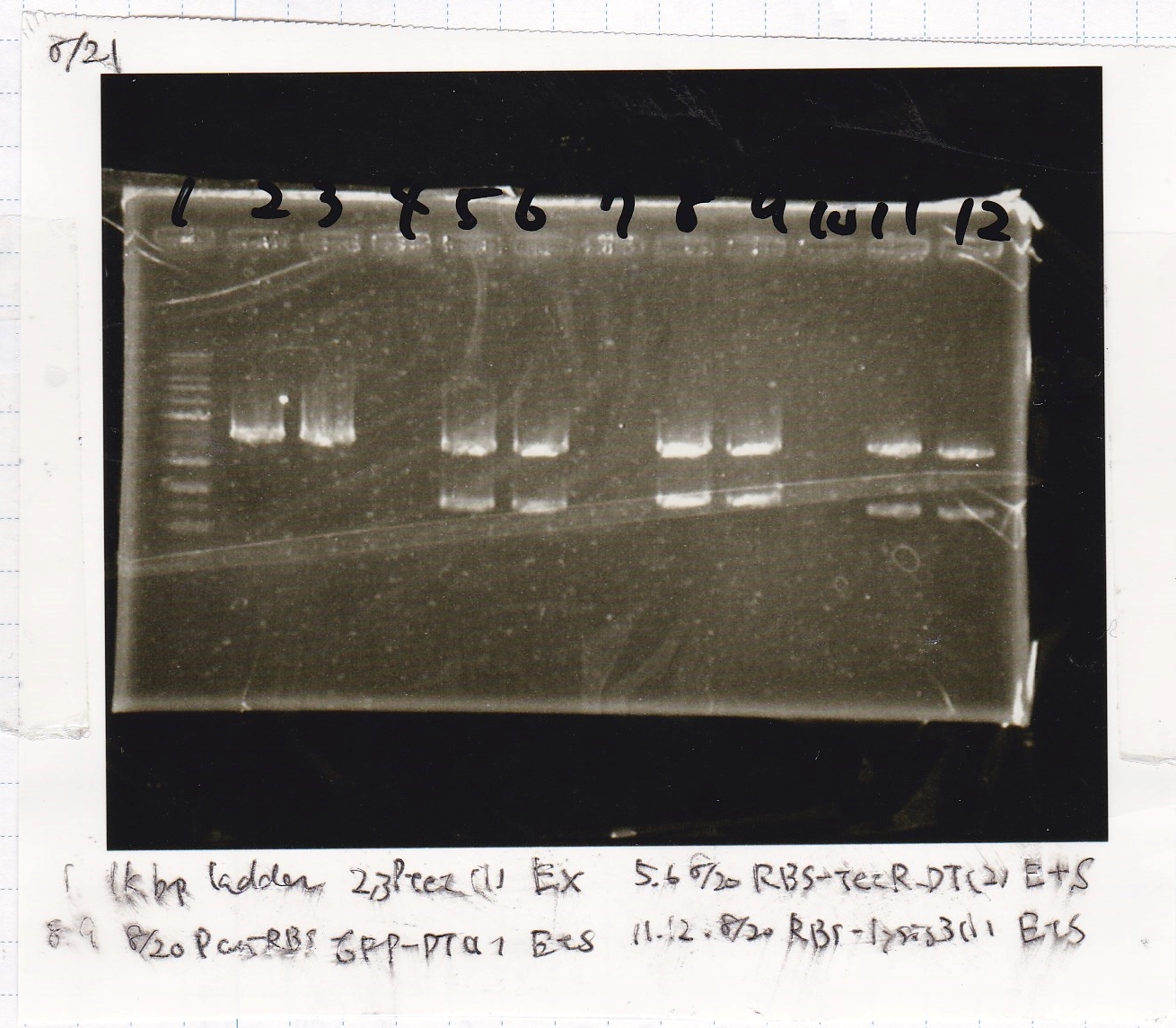
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kbp ladder | -- |
| 2 | 8/20 Ptet -(1) | EcoRI & XbaI |
| 3 | ||
| 5 | 8/20 RBS-tetR-DT -(2) | EcoRI & SpeI |
| 6 | ||
| 8 | 8/20 Pconst-RBS-GFP-DT -(1) | EcoRI & SpeI |
| 9 | ||
| 11 | 8/20 RBS-lysis3 -(1) | EcoRI & SpeI |
| 12 |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Ptet (EcoRI & XbaI) | 21 | 1.55 | 0.78 |
| RBS-tetR-DT (EcoRI & SpeI) | 12 | 1.45 | 0.62 |
| Pconst-RBS-GFP-DT (EcoRI & SpeI) | 11 | 1.36 | 0.60 |
| RBS-lysis3 (EcoRI & SpeI) | 10 | 1.26 | 0.50 |
Ristriction Enzyme Digestion
| 8/20 Ptet-(1) | EcoRI | XbaI | 10x BSA | 10x buffer M | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 4.5 | 1 | 1 | 3 | 3 | 17.5 | 30 |
| 1 cut | 0.5 | 0.2 | 0 | 1 | 1 | 7.3 | 10 |
| 1 cut | 0.5 | 0 | 0.2 | 1 | 1 | 7.3 | 10 |
| NC | 0.5 | 0 | 0 | 1 | 1 | 7.5 | 10 |
| 8/20 Pconst-RBS-luxR-DT-(2) | EcoRI | XbaI | 10x BSA | 10x buffer M | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 2.9 | 1 | 1 | 3 | 3 | 19.1 | 30 |
| 1 cut | 0.3 | 0.2 | 0 | 1 | 1 | 7.5 | 10 |
| 1 cut | 0.3 | 0 | 0.2 | 1 | 1 | 7.5 | 10 |
| NC | 0.3 | 0 | 0 | 1 | 1 | 7.7 | 10 |
| 8/17 DT | EcoRI | XbaI | 10x BSA | 10x buffer M | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 5.3 | 1 | 1 | 3 | 3 | 16.7 | 30 |
| 1 cut | 0.5 | 0.2 | 0 | 1 | 1 | 7.3 | 10 |
| 1 cut | 0.5 | 0 | 0.2 | 1 | 1 | 7.3 | 10 |
| NC | 0.5 | 0 | 0 | 1 | 1 | 7.5 | 10 |
- incubate at 37°C for 1h
Electrophoresis
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | 1kbp ladder | -- | -- |
| 2 | 8/20 Ptet -(1) | EcoRI | XbaI |
| 3 | 8/20 Ptet -(1) | EcoRI | -- |
| 4 | 8/20 Ptet -(1) | -- | XbaI |
| 5 | 8/20 Ptet -(1) | -- | -- |
| 6 | 8/20 J23100-RBS-luxR-DT -(2) | EcoRI | XbaI |
| 7 | 8/20 J23100-RBS-luxR-DT -(2) | EcoRI | -- |
| 8 | 8/20 J23100-RBS-luxR-DT -(2) | -- | XbaI |
| 9 | 8/20 J23100-RBS-luxR-DT -(2) | -- | -- |
| 10 | 8/20 DT | EcoRI | XbaI |
| 11 | 8/20 DT | EcoRI | -- |
| 12 | 8/20 DT | -- | XbaI |
| 13 | 8/20 DT | -- | -- |
| 14 | 1kbp ladder | -- | -- |
| 15 | 8/20 Ptet -(1) | EcoRI | XbaI |
- The reason why lane 2 and 15 were same is that the well of lane 2 might be broken.
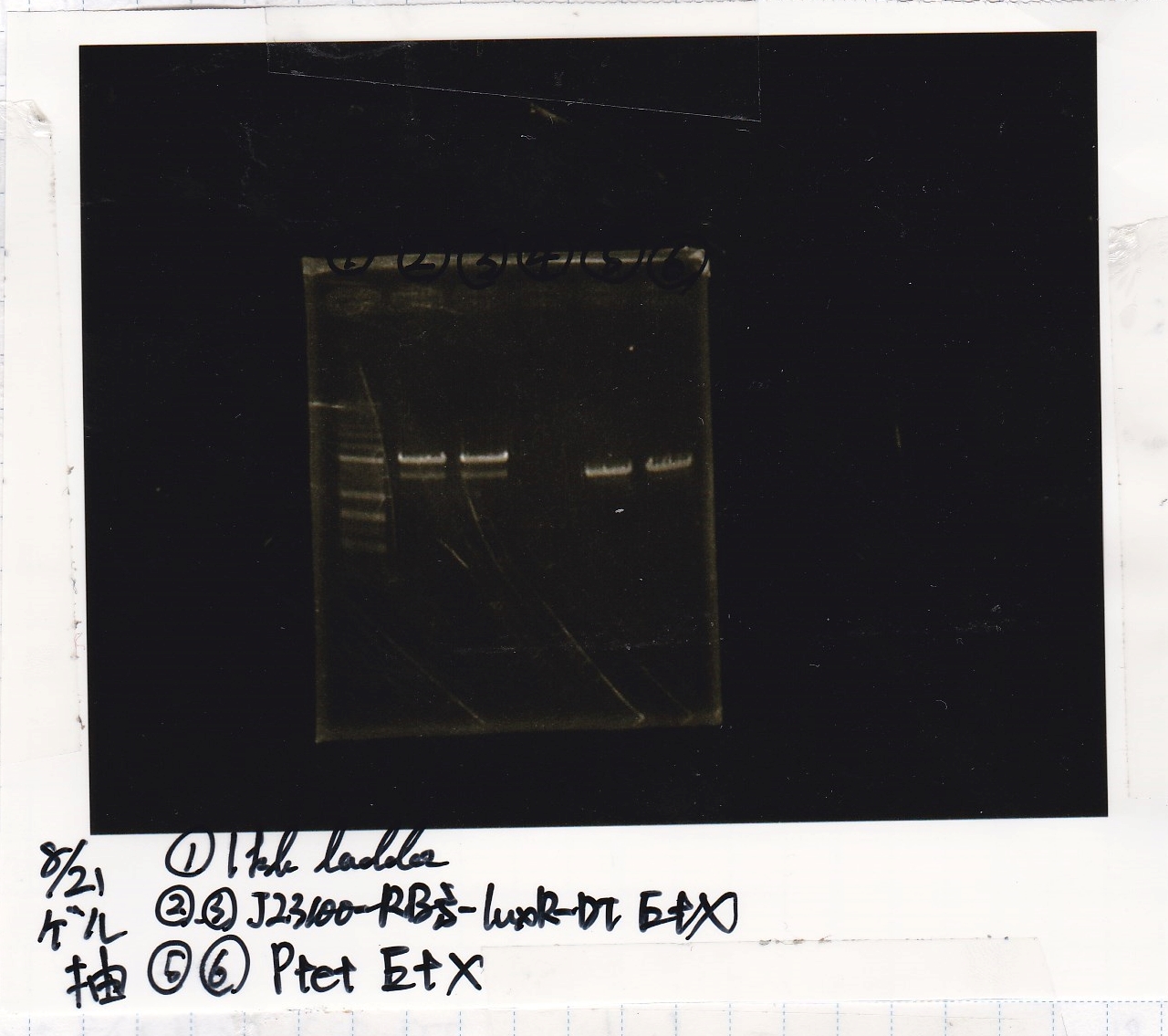
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | 1kbp ladder | -- | -- |
| 2 | 8/20 J23100-RBS-luxR-DT -(2) | EcoRI | XbaI |
| 3 | 8/20 DT | EcoRI | XbaI |
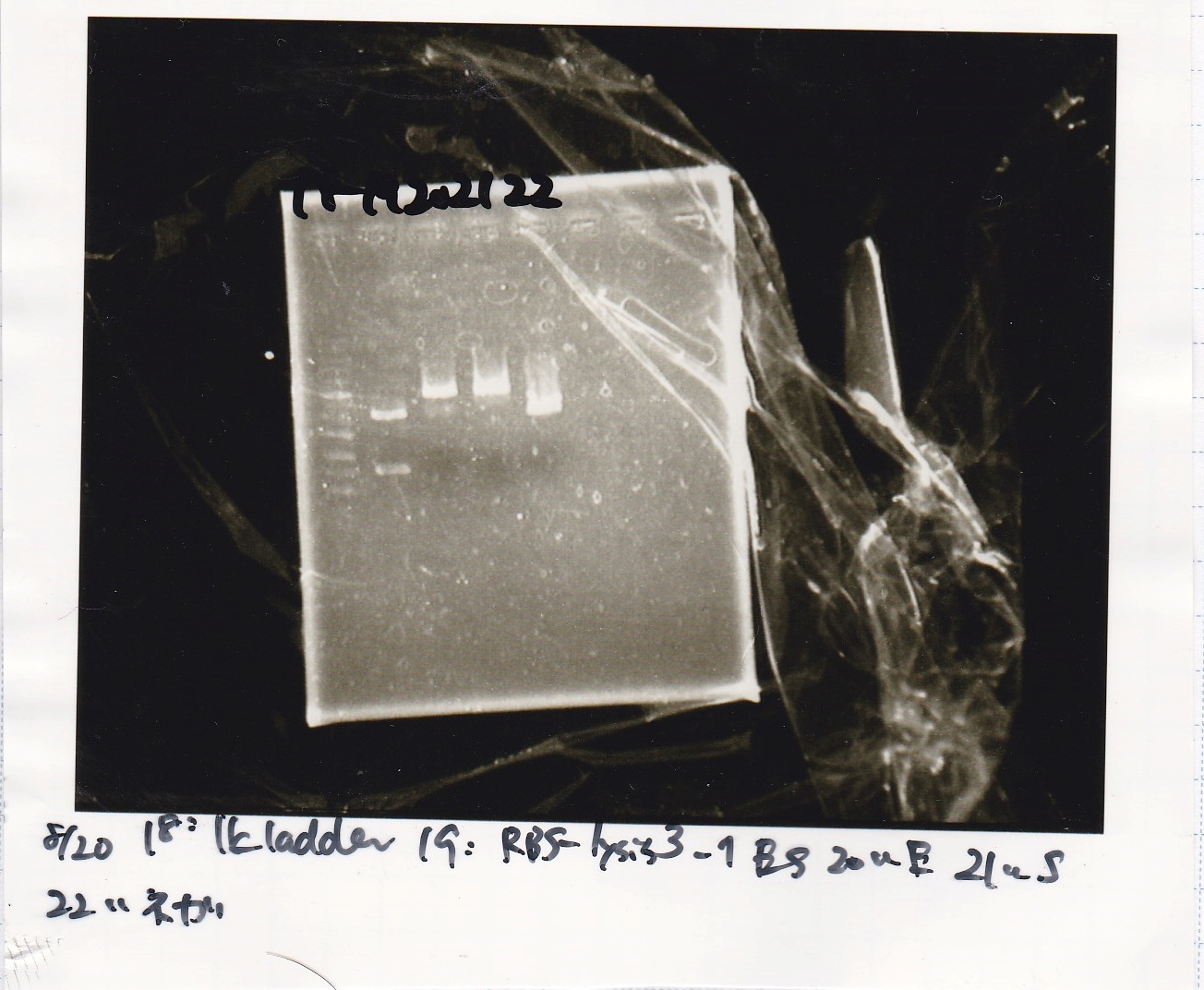
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kb ladder | -- |
| 2 | 8/21 J23100-RBS-luxR-DT 25µL | EcoRI+XbaI |
| 3 | 8/21 J23100-RBS-luxR-DT 25µL | EcoRI+XbaI |
| 4 | -- | -- |
| 5 | 8/21 Ptet 25µL | EcoRI+XbaI |
| 6 | 8/21 Ptet 25µL | EcoRI+XbaI |
Electrophoresis
Ligation
| state | Vector | Inserter | Ligation High ver.2 | total | ||
|---|---|---|---|---|---|---|
| experiment | 8/21 Pcn-RBS-luxR-DT(+Amp) | 2.3 | 8/21 Pcn-RBS-GFP-DT | 1.7 | 2 | 6 |
| NC | 8/21 Pcn-RBS-luxR-DT(+Amp) | 2.3 | MilliQ | 1.7 | 2 | 6 |
| experiment | 8/18 Plux(+CP) | 1.1 | 8/19 RBS-GFP-DT | 5 | 3.1 | 9.2 |
| NC | 8/18 Plux(+CP) | 1.1 | MilliQ | 5 | 3.1 | 9.2 |
| experiment | 8/18 RBS(+Amp) | 2.2 | 8/19 lysis1 | 6.0 | 4.1 | 12.3 |
| experiment | 8/18 RBS(+Amp) | 2.2 | 8/19 lysis2 | 3.6 | 2.9 | 8.7 |
| NC | 8/18 RBS(+Amp) | 2.2 | MilliQ | 3.6 | 2.9 | 8.7 |
| experiment | 8/21 Ptet | 1.5 | 8/20 RBS-tetR-DT (2) | 2.1 | 1.8 | 5.4 |
| NC | 8/21 Ptet | 1.5 | MilliQ | 2.1 | 1.8 | 5.4 |
Transformation
| Name | Sample | Competent Cells | Total | Plate |
|---|---|---|---|---|
| 8/21 Pcon-RBS-GFP-DT+Pcon-RBS-GFP-DT | 1µL | 10µL | 11µL | Amp |
| 8/21 Pcon-RBS-GFP-DT NC | 1µL | 10µL | 11µL | Amp |
| 8/18 RBS+8/19 lysis1 | 1µL | 10µL | 11µL | Amp |
| 8/18 RBS+8/19 lysis2 | 1µL | 10µL | 11µL | Amp |
| 8/18 RBS NC | 1µL | 10µL | 11µL | Amp |
| 8/18 Plux+8/18 RBS-GFP-DT | 1µL | 10µL | 11µL | CP |
| 8/18 Plux+8/18 RBS-GFP-DT | 1µL | 10µL | 11µL | CP |
| 8/21 Ptet(pm)+8/20 RBS-tetR-DT(2) | 1µL | 10µL | 11µL | CP |
| 8/21 Ptet(pm)+8/20 RBS-tetR-DT(2) NC | 1µL | 10µL | 11µL | CP |
| 8/21 pSB1C3 | 1µL | 10µL | 11µL | CP |
LB Medium Plate
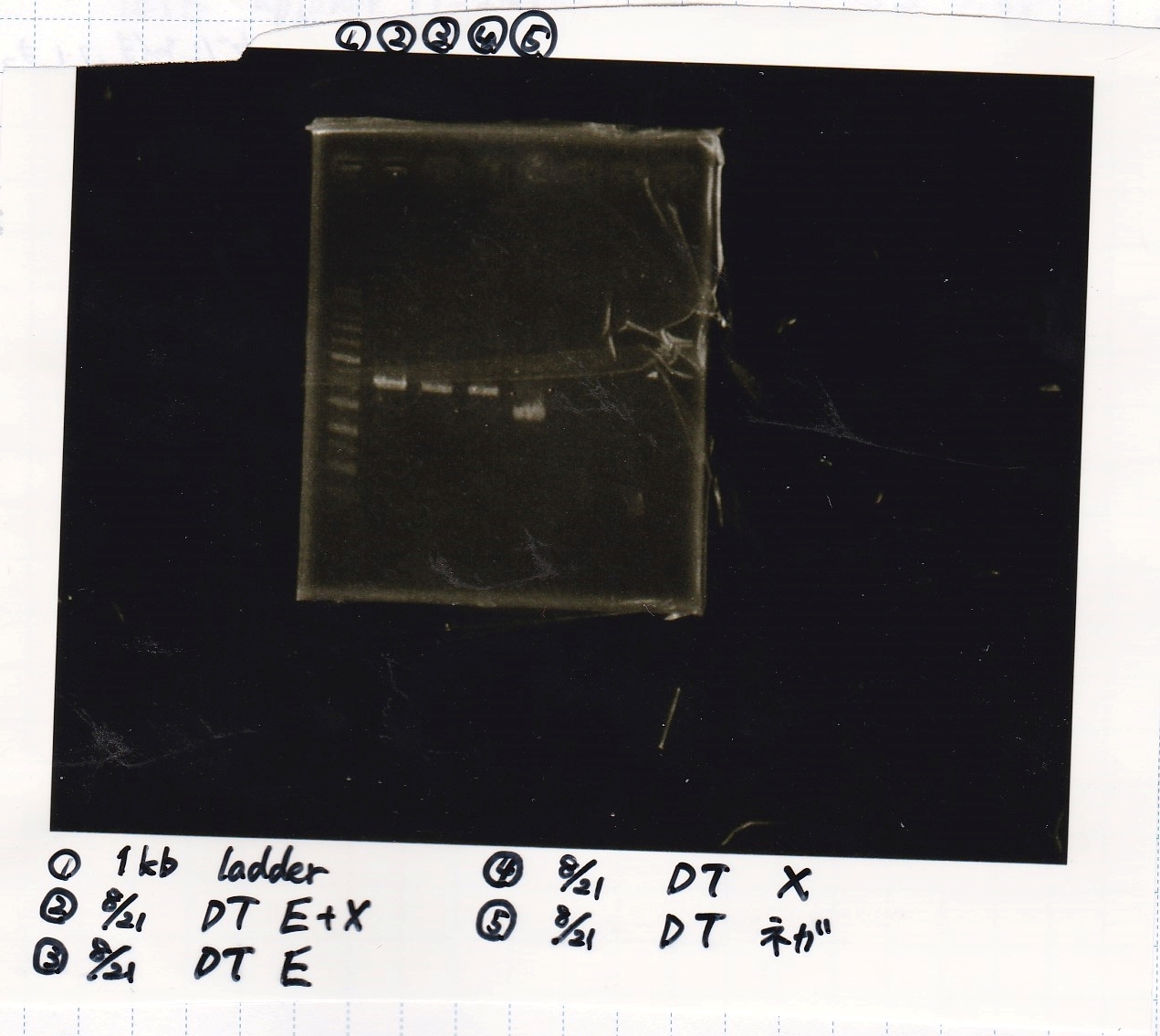
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kb ladder | -- |
| 2 | -- | -- |
| 3 | 8/21 DT | EcoRI & XbaI |
| 4 |
File:Igku Aug21 Gel Extraction(DT EcoRI+XbaI 12,5μL①)before.jpg
File:Igku Aug21 Gel Extraction(DT EcoRI+XbaI 12,5μL②)after.jpg
Miniprep
| DNA | concentration [µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| tRNA-spinach-(1) | 234 | 1.64 | 2.14 |
| tRNA-spinach-(2) | 440 | 1.60 | 1.91 |
| tetR-aptamer 12-p(1076)-(1) | 150 | 1.61 | 2.03 |
| tetR-aptamer 12-p(1076)-(2) | 374 | 1.45 | 1.66 |
| tetR-aptamer 12-1R(113)-(1) | 304 | 1.68 | 2.21 |
| tetR-aptamer 12-1R(113)-(2) | 382 | 1.47 | 1.66 |
| tetR-aptamer 12-1M(105)-(1) | 374 | 1.68 | 2.20 |
| tetR-aptamer 12-1M(105)-(2) | 286 | 1.31 | 1.43 |
| PT181 attenuator-(1) | 330 | 1.66 | 2.11 |
| PT181 attenuator-(2) | 270 | 1.68 | 2.18 |
| Fusion1 attenuator-(1) | 306 | 1.67 | 2.20 |
| Fusion1 attenuator-(2) | 282 | 1.60 | 1.94 |
| Fusion3m2 attenuator-(1) | 332 | 1.68 | 2.25 |
| Fusion3m2 attenuator-(2) | 326 | 1.67 | 2.10 |
| PT181 antisense-(1) | 178 | 1.64 | 1.68 |
| PT181 antisense-(2) | 116 | 1.63 | 1.95 |
| Fusion1 antisense-(1) | 300 | 1.67 | 2.21 |
| Fusion1 antisense-(2) | 164 | 1.42 | 1.46 |
| Fusion6 antisense-(1) | 336 | 1.65 | 2.18 |
| Fusion6 antisense-(2) | 192 | 1.65 | 2.01 |
 "
"