From 2013.igem.org
(Difference between revisions)
|
|
| Line 281: |
Line 281: |
| | |12||1kbp ladder||-- | | |12||1kbp ladder||-- |
| | |} | | |} |
| - | [[File:igku_xxbeforexx.xxx]]<br> | + | [[File:igku_9105.jpg]]<br> |
| - | [[File:igku_xxafterxx.xxx]]<br> | + | [[File:igku_9106.jpg]]<br> |
| | | | |
| | </div> | | </div> |
Revision as of 07:08, 26 September 2013
Sep 10
Ligation
Nakamoto
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/6 Pcon-RBS-luxR-DT | 1.9µL | 9/6Plux-RBS-GFP-DT | 22 µL | µL
|
| experiment | 9/6 Plux | 2µL | 9/8 RBS-lysis1-DT | 6.6µL | xx µL
|
| experiment | 9/6 Plux | 2µL | 9/8 RBS-lysis2-DT | 6.3µL | xx µL
|
| experiment | 9/8 Ptet-PT181 antisense | 1.6µL | 9/8 spinach-DT | 4.4µL | xx µL
|
| experiment | 9/8 Pcon-PT181 attenuator | 3.4µL | 9/8 apz 12-1R-PT | 2.1µL | xx µL
|
| experiment | 9/8 Pcon | 3.4µL | 9/8 apz 12-1R-PT | 2.3µL | xx µL
|
| experiment | 9/8 DT | 3.0µL | 9/8 Pcon-PT181 attenuator | 7.9µL | xx µL
|
| experiment | 9/10 pSB1C3 | 1.8µL | 9/8 F3m2 attenuator | 4.6µL | xx µL
|
| experiment | 9/10 pSB1C3 | 1.8µL | 9/8 F1 attenuator | 3.4µL | xx µL
|
Miniprep
Nakamoto
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| spinach-DT | 202.4 | 1.97 | 1.93
|
| Plux-RBS-GFP-DT | 164.2 | 2.80 | 2.06
|
| Pcon-DT attenuator | 205.4 | 1.81 | 2.39
|
| RSBJC3-2 | 240.8 | 1.73 | 1.43
|
| Plaz-1 | 240.8 | 1.73 | 1.43
|
Restriction Enzyme Digestion
Nakamoto
| | 9/10 Plux-RBS-GFP-DT | EcoR1 | Spel | Buffer | BSA | MilliQ | total
|
| 2cuts | 12.1µL | 1µL | 1µL | 3µL | 3µL | 9.9µL | 30µL
|
| NC | 0.6µL | 0µL | 0µL | 1µL | 1µL | 7.4µL | 10µL
|
| | 9/10 Spinach-DT | Xbal | Pstl | Buffer | BSA | MilliQ | total
|
| 2cuts | 9.9µL | 1µL | 1µL | 3µL | 3µL | 12.1µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| | 9/6 apt12-1R -DT | Xbal | Pstl | Buffer | BSA | MilliQ | total
|
| 2cuts | 8.0µL | 1µL | 1µL | 3µL | 3µL | 14.0µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 9/10 Pcon-pt81 attenuator | EcoR1 | Spel | Buffer | BSA | MilliQ | total
|
| 2cuts | 9.8µL | 1µL | 1µL | 3µL | 3µL | 12.2µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Plux-RBS-GEP-DT | EcoR1 | Spel
|
| 3 | Plux-RBS-GEP-DT | -- | --
|
| 4 | Spinach-DT | Xbal | Pstl
|
| 5 | Spinach-DT | -- | --
|
| 6 | aptamer 121R-DT | Xbal | Pstl
|
| 7 | aptamer 121R-DT | -- | --
|
| 8 | Pcon-pt181 attenuator | EcoR1 | Spel
|
| 9 | Pcon-pt181 attenuator | -- | --
|
| 10 | 100bp ladder | -- | --
|

Colony PCR
Tatsui
| Sample | base pair
|
| 9/9 Pcon-pT181 antisense | --
|
| 9/8 J223100(entA) | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min10sec | 30
|
Electrophoresis
Tatsui
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Pcon Pt181 antisense1+spinach-DT
|
| 3 | Pcon Pt181 antisense2+spinach-DT
|
| 4 | Pcon Pt181 antisense3+spinach-DT
|
| 5 | Pcon Pt181 antisense4+spinach-DT
|
| 6 | Pcon Pt181 antisense5+spinach-DT
|
| 7 | Pcon Pt181 antisense6+spinach-DT
|
| 8 | Pcon Pt181 antisense7+spinach-DT
|
| 9 | Pcon Pt181 antisense8+spinach-DT
|
| 10 | Pcon Pt181 antisense9+spinach-DT
|
| 11 | Pcon Pt181 antisense10+spinach-DT
|
| 12 | Pcon Pt181 antisense11+spinach-DT
|
| 13 | Pcon Pt181 antisense12+spinach-DT
|
| 14 | 100bp ladder
|
File:Igku xxxxxx.xxx
| Lane | Sample
|
| 1 | 1kbp ladder
|
| 2 | Pcon entA1
|
| 3 | Pcon entA2
|
| 4 | Pcon entA3
|
| 5 | 1kbp ladder
|
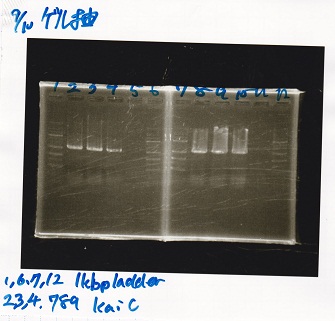
Tatsui
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 2 | KaiA(200pg) | --
|
| 3 | KaiA(200pg) | --
|
| 4 | -- | --
|
| 5 | KaiA(400pg) | --
|
| 6 | KaiA(400pg) | --
|
| 7 | -- | --
|
| 8 | KaiA(800pg | --
|
| 9 | KaiA(800pg) | --
|
| 10 | 1kbp ladder | --
|


| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 2 | PKaiBC(200pg) | --
|
| 3 | PKaiBC(200pg) | --
|
| 4 | -- | --
|
| 5 | PKaiBC(400pg) | --
|
| 6 | PKaiBC(400pg) | --
|
| 7 | -- | --
|
| 8 | PKaiBC(800pg | --
|
| 9 | PKaiBC(800pg) | --
|
| 10 | 1kbp ladder | --
|
Liquid Culture
Hirano
| Sample | medium
|
| 9/4 J23100①(JM109) | LB(Amp)
|
| 9/8 J23100① (entA) | LB(Amp)
|
Tatsui
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 2 | KaiC | --
|
| 3 | KaiC | --
|
| 4 | KaiC | --
|
| 5 | -- | --
|
| 6 | 1kbp ladder | --
|
| 7 | 1kbp ladder | --
|
| 8 | KaiC | --
|
| 9 | KaiC | --
|
| 10 | KaiC | --
|
| 11 | -- | --
|
| 12 | 1kbp ladder | --
|

Liquid Culture
Tatsui
| Sample | medium
|
| 9/10 Pcon-pt181 antisense-Spinach-DT | Plusgrow
|
Colony PCR
No name
| Sample | base pair
|
| 9/9 Phad larac-1 RBS-lux1-DT | 2331
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 2min | 30
|
| Sample | base pair
|
| 9/9 Pcon-lacZα (psB4KS) | 712
|
| 9/9 Pcon-Spinach-DT(psB4KS) | 605
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 42s | 30
|
| Sample | base pair
|
| 9/9 Plac+RBS-lysis3}-DT | 1273
|
| 9/9 Pcon+RBS-lux1-DT | 1079
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min12s | 30
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | 9/9 Pcou-RBS-lacZα-DT(p3B4KS9)-1 | -- | --
|
| 3 | 9/9 Pcou-RBS-lacZα-DT(p3B4KS9)-2 | -- | --
|
| 4 | 9/9 Pcou-Spinach-DT(pB4KS)-1 | -- | --
|
| 5 | 9/9 Pcou-Spinach-DT(pB4KS)-2 | -- | --
|
| 6 | 100bp ladder | -- | --
|
File:Igku xxxxxx.xxx
Transformation
Nakamoto
| Name | Sample | Competent Cells | Total | Plate
|
| F1 attenuator +p3B1C3 | µL | µL | µL | CP
|
| F3n2 attenuator +p3B1C3 | µL | µL | µL | CP
|
| F1 antisense +p3B1C3 | µL | µL | µL | CP
|
| F6 antisense+p3B1C3 | µL | µL | µL | CP
|
| apt12-P +p3B1C3 | µL | µL | µL | CP
|
| apt12-1M +p3B1C3 | µL | µL | µL | CP
|
| Pcon-luxR+Plux-GFP | µL | µL | µL | Amp
|
| Plux+RBS-lysis1-DT | µL | µL | µL | CP
|
| Plux+RBS-lysis2-DT | µL | µL | µL | CP
|
| Ptet-pT181antisense+Spinach-DT | µL | µL | µL | CP
|
| Pcon-pT181 attenuator+apt121R-DT | µL | µL | µL | Amp
|
| Pcon-1 apt12-1R-DT | µL | µL | µL | Amp
|
| DT+Pcon-pT181 attenuator | µL | µL | µL | CP
|
Electrophoresis
Okazaki
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Spinach-DT | Xbal | Ptsl
|
| 3 | Spinach-DT | Xbal | Ptsl
|
| 4 | Spinach-DT | Xbal | Ptsl
|
| 5 | Spinach-DT | Xbal | Ptsl
|
| 6 | 100bp ladder | -- | --
|
File:Igku xxxxxx.xxx
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Pcon-pT181 attenuator | EcoR1 | Spel
|
| 3 | Pcon-pT181 attenuator | EcoR1 | Spel
|
| 4 | Pcon-pT181 attenuator | EcoR1 | Spel
|
| 5 | Pcon-pT181 attenuator | EcoR1 | Spel
|
| 6 | 100bp ladder | -- | --
|
File:Igku xxxxxx.xxx
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | Plux-RBS-GFP-DT | EcoR1 | Spel
|
| 2 | Plux-RBS-GFP-DT | EcoR1 | Spel
|
| 3 | Plux-RBS-GFP-DT | EcoR1 | Spel
|
| 4 | Plux-RBS-GFP-DT | EcoR1 | Spel
|
| 5 | 100bp ladder | -- | --
|
| 6 | apt12-1R-DT | Xbal | Pstl
|
| 7 | apt12-1R-DT | Xbal | Pstl
|
| 8 | apt12-1R-DT | Xbal | Pstl
|
| 9 | apt12-1R-DT | Xbal | Pstl
|
File:Igku xxxxxx.xxx
 "
"




