Team:Greensboro-Austin/Smell degradation
From 2013.igem.org
Ryanlannan (Talk | contribs) (→References) |
Ryanlannan (Talk | contribs) (→References) |
||
| Line 43: | Line 43: | ||
=References= | =References= | ||
| - | + | # Kasai, D. et al. Uncovering the Protocatechuate 2,3-Cleavage Pathway Genes. Journal of Bacteriology 191, 6758–68 (2009). | |
Revision as of 18:33, 25 June 2013
Contents |
Introduction
p-Cresol is introduced into the environment from a number of natural and anthropogenic sources. As p-cresol is toxic and malodorous, industries producing p-cresol have sought ways to reduce its emission in order to limit their impact on water and air quality. The swine industry is one of the most concerned about the problem of p-cresol pollution. This compound is a primary constituent of swine manure odor as a result of gut flora degradation of tyrosine. More recently, with the introduction of concentrated animal feeding operations (CAFOs), the amount of pig fecal matter being produced is significant, causing complaints from nearby communities. If the malodorous nature of this waste could be reduced by lowering the p-cresol content, it would be of immediate benefit to these areas.
Pseudomonas putida
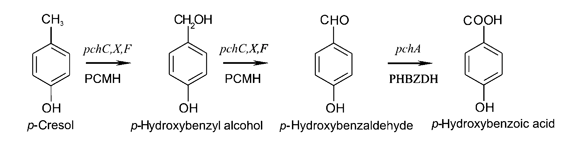
Strains of Pseudomonas putida that are able to naturally degrade aromatic compounds, including p-cresol, have been discovered and a few have been characterized. These frequently occur in water sources polluted with aromatic compounds, such as runoff from coal refineries. The strain NCIMB 9866 contains the pchACXF gene cluster, capable of converting p-cresol to 4-hydroxybenzoic acid.
Bacillus sp.
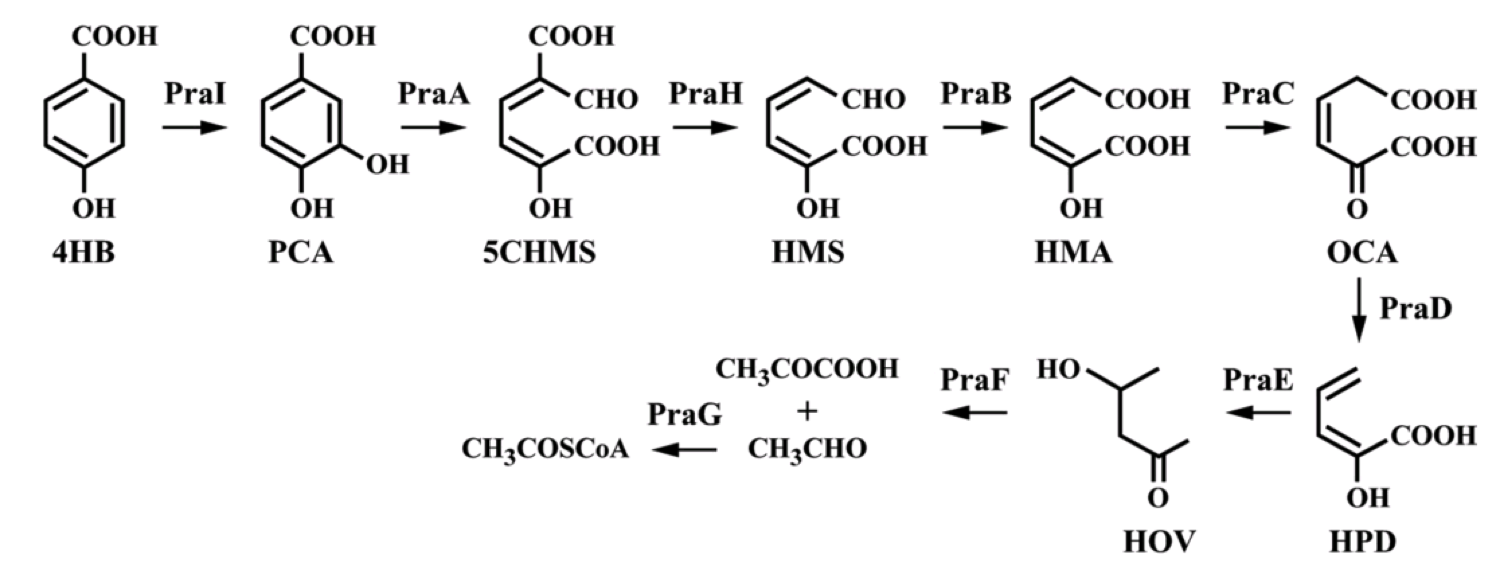
Genes with the ability to metabolize 4-hydroxybenzoic acid were identified and characterized in Bacillus sp. These genes were subsequently expressed in E. coli, which then had the ability to grow on 4-hydroxybenzoic acid as a sole carbon source (Kasai, et al., 2009). The Bacillus sp. strain JJ-1b contains this praABEGFDCHI gene cluster, capable of converting 4-hydroxybenzoic acid to pyruvate and Acetyl-CoA.
Strategy
Escherichia coli
The aforementioned gene clusters pchACXF and praABEGFDCHI will be cloned into separate plasmids and transformed into the single-gene knock-out strain of BW25113 E. coli that has had the gene coding for tryptophanase removed. Tryptophanase is an enzyme that converts tryptophan into indole, the compound responsible for the characteristic smell of E. coli. Without the ability to produce indole from tryptophan, the strain itself is effectively odorless.
Directed Evolution
In addition, the untransformed knock-out strain of BW25113 E. coli will undergo directed evolution to select for reduction of p-cresol toxicity.
References
- Kasai, D. et al. Uncovering the Protocatechuate 2,3-Cleavage Pathway Genes. Journal of Bacteriology 191, 6758–68 (2009).
 "
"